Construction and identification of genomic expression library of Streptococcus agalactiae virulent strain Hod-1 isolated from Oreochromis niloticus
-
摘要:
为了筛选罗非鱼(Oreochromis niloticus)无乳链球菌(Streptococcus agalactiae)在鱼体内特异诱导表达基因,该研究构建了罗非鱼无乳链球菌强毒株Hod-1的基因组表达文库。采用Sau3AⅠ对无乳链球菌菌株Hod-1基因组DNA进行不完全酶切,回收大小为0.5~2.0 kb的DNA片段,并与经BamHⅠ单酶切且去磷酸化的pET-28 a/b/c表达载体连接,转化表达宿主菌BL21(DE3)。结果显示,所构建的无乳链球菌菌株Hod-1基因组表达文库含1.024×105个克隆,远大于覆盖无乳链球菌全基因组所需的最小库容量(45 579个)。PCR检测结果显示重组子中外源片段的插入率为94%,插入片段大小为0.5~2.0 kb,且分布均匀,测序结果显示插入序列与GenBank中无乳链球菌基因组序列同源性高达99%以上。结果表明该研究成功构建了罗非鱼无乳链球菌的基因组表达文库,该文库可用于无乳链球菌在罗非鱼体内诱导表达基因的筛选。
Abstract:To screen the specific genes of Streptococcus agalactiae induced in Oreochromis niloticus, the genomic expression library of the virulent strain Hod-1 was constructed. The Sau3AⅠ was used to digest genomic DNA of S.agalactiae strain Hod-1 incompletely. The DNA fragments ranging from 0.5 kb to 2 kb were recycled and ligated into pET-28 a/b/c expression vectors which had been digested by BamHⅠ and dephosphorylated by SAP before using. The recombinant vectors were transformed into BL21 (DE3). The results show that the genomic expression library of S.agalactiae contained 1.024×105 clones, considerably more than the minimum number of clones (45 579 clones) covering the whole genomic DNA of S.agalactiae. Results of PCR detection show that the rate of fragment insertion was 94% and different sizes of inserted fragments ranged from 0.5 kb to 2.0 kb. The results of sequencing show that the homology between inserted sequences and the genomic DNA sequnce of S.agalactiae was over 99%. In conclusion, the genomic expression library of S.agalactiae strain Hod-1 was constructed successfully, which can be used for screening genes of S.agalactiae induced expression in O.niloticus.
-
浮游植物是海洋生态系统中最基础的一个环节,作为该系统最重要的初级生产者,在海洋生态系统的物质循环和能量流动中起着重要的作用。浮游植物的种类组成和数量变动都将直接或者间接地制约海洋生产力的发展,从而影响海洋生物资源的开发和利用[1-5]。珠江口南沙海域位于广州市最南端,濒珠江出海口伶仃洋,属咸淡水的交汇区域,同时受到地表径流和海洋潮汐的影响,生态环境复杂,形成独特的海洋生物群落。近年来,随着周边地区经济社会的快速发展,以及滩涂围垦、航道疏浚和养殖等人类活动规模不断扩大,使得海水的富营养化加剧,极大地影响了该海区生态群落结构的稳定性,增大了发生赤潮的危险性,珠江口海域已成为中国赤潮的多发区之一[6-7]。
目前,对珠江河口及近海浮游生物的群落特征的研究已有报道[8-12],但是对珠江口内海域研究的比较少,特别是人为扰动最为频繁的珠江口南沙区周围的海域。文章通过对2015年4月(丰水期)和11月(枯水期)珠江口南沙海域生态综合调查采集的浮游植物样品进行鉴定分析,初步研究了该海域海洋浮游植物的种类组成、优势种、细胞丰度的分布及多样性水平,旨在为研究该海域生产力水平、海洋生态系统的群落结构和环境质量状况提供基础数据。
1. 材料与方法
1.1 采样和分析
2015年4月(丰水期)和11月(枯水期)在珠江口南沙区周围的海域(113°29′52″E~113°47′19″E,22°28′10″N~22°51′23″N)进行浮游植物样品采集,丰水期和枯水期都布设了20个调查站位(S1~S20,图 1)。调查站点均位于珠江口南沙海域人类活动扰动频繁海域,覆盖了从近口淡水、咸淡水段到口外海水段的河口典型生境特征。浮游植物样品用浅水Ⅲ型浮游生物网自底至表垂直拖网采集,并当场加入甲醛溶液保存。带回实验室后浓缩至50 mL,于显微镜下进行鉴定与计数,测定分析其种类组成、数量、分布、优势度、多样性指数和均匀度等。
使用YSI便携式温盐深仪现场采集各站位盐度、温度、溶解氧(DO)、pH和水深等物理环境数据;同时现场采集海水样品,带回实验室后分析水体中的溶解性无机氮(TIN)、活性磷酸盐(PO4-P)和叶绿素a(Chl-a)的含量。浮游植物样品的采集和分析均按《海洋监测规范》(GB17378—2007)和《海洋调查规范-海洋生物调查》(GB12763.6—2007)[13-14]中规定的方法进行。
1.2 评价方法
浮游植物优势度(Y)应用以下公式计算:
$ Y = \frac{{{n_i}}}{N} \times {f_i} $
式中ni为第i种的个体数;fi是该种在各站中出现的频率;N为所有站每个种出现的总个体数,以Y>0.02作为优势种。
采用Shannon-Wiener[15]、Pielou[16]测定浮游植物的多样性指数(H′)、均匀度指数(J′),其计算公式分别为:
$ \begin{array}{l} H' =-\sum\limits_{i = 1}^S {{P_i}{\rm{lo}}{{\rm{g}}_2}{P_i}} \\ J' = \frac{{H'}}{{{\rm{lo}}{{\rm{g}}_2}S}} \end{array} $
式中S为样品中的种类总数;Pi为第i种的个体数与总个体数的比值。
浮游植物与环境因子的相关性分析采用SPSS 19.0统计软件分析;典范对应分析(canonical correspondence analysis,CCA)应用CANOCO 4.0软件进行,排序结果用物种-环境因子关系的双序图表示。另外,采用孙军等[17]的浮游植物多样性的评判阈值(表 1)对浮游植物多样性进行等级评价。
表 1 浮游植物多样性指标的评判阈值Table 1. Threshold value evaluation for diversity indices of phytoplankton多样性指标
diversity index很低
very low
(Ⅰ)低
low
(Ⅱ)一般
normal
(Ⅲ)高
high
(Ⅳ)很高
very high
(Ⅴ)种类数species number <15 15~24 25~34 35~44 >45 多样性指数(H′)Shannon-Wiener diversity index <1.5 1.5~2.49 2.5~3.49 3.5~4.5 >4.5 均匀度指数(J′)Pielous evenness index <0.68 0.68~0.73 0.74~0.79 0.80~0.88 >0.88 2. 结果与分析
2.1 浮游植物物种组成
共鉴定出浮游植物44个属76种,隶属于硅藻门、甲藻门、绿藻门、蓝藻门和裸藻门5大门类。其中丰水期共鉴定出浮游植物5门37属68种(包括变种和变形),硅藻门种类最多(24属39种),占总种数的57.35%;其次是绿藻门(7属15种),占总种数的22.06%;甲藻门有2属7种,占总种数的10.29%;裸藻门有3属5种,占总种数的7.35%;蓝藻门有1属2种,占总种数的2.94%。S1站鉴定出的浮游植物种类最多(38种),而S17站鉴定出的浮游植物种类最少(6种),但都是属于硅藻类。各站位中新月筒柱藻(Cylindrotheca closterium)、中肋骨条藻(Skeletonema costatum)、虹彩圆筛藻(Coscinodiscus oculusiridis)在每个站位中都有分布。此外,奇异棍形藻(Bacillaria paradoxa)也分布比较广泛(表 2)。
表 2 珠江口南沙海域浮游植物物种组成Table 2. Species composition of phytoplankton in the Nansha sea area of Pearl River Estuary门类
phylum总种类数
total species number丰水期(5月)
wet season (May)枯水期(11月)
dry season (November)种类
species比例/%
percentage种类
species比例/%
percentage种类
species比例/%
percentage硅藻Bacillariophyta 39 51.32 39 57.35 33 55.00 甲藻Pyrrophyta 15 19.74 7 10.29 14 23.33 绿藻Chlorophyta 15 19.74 15 22.06 8 13.33 蓝藻Cyanophyta 2 2.63 2 2.94 2 3.33 裸藻Euglenophyta 5 6.58 5 7.35 3 5.00 总计total 76 - 68 - 60 - 枯水期共鉴定出浮游植物5门44个属60种(包括变种和变形),其中还是硅藻门种类最多(22属33种),占总种数的55.00%;其次是甲藻门(11属14种),占总种数的23.33%;绿藻门有6属8种,占总种数的13.33%;裸藻门有3属3种,占总种数的5.00%;蓝藻门有2属2种,占总种数的3.33%。中肋骨条藻、虹彩圆筛藻、有棘圆筛藻(C.spinosus)、布氏双尾藻(Ditylum brightwelli)在大部分调查站位中均有分布(表 2)。可见珠江口南沙海域丰水期和枯水期的浮游植物物种组成相似,都是硅藻类占主体。
2.2 浮游植物种类的分布
浮游植物的区域分布具有季节差异,以丰水期较明显。丰水期S1站和S2站分别有38和30种,而S17站只有6种,表现为由河口内向外海递减的特征。枯水期该水域各站位浮游植物种类数比较接近(12~16种),也表现为由河口内向外海递减的特征(图 2)。
2.3 浮游植物细胞丰度的变化及分布
丰水期调查结果表明(图 3),浮游植物细胞丰度为(361.52~2 000.73)×104个· L-1,平均为651.91×104个· L-1,主要集中在万顷沙旁的水道,以S17站最高,其次是S19站(912.59×104个·L-1),S14站最低。细胞丰度组成大部分以硅藻为主,硅藻类平均细胞丰度为636.46×104个· L-1,占该海域浮游植物平均细胞丰度的97.63%,不同站位硅藻细胞丰度占比范围为92.95%~100.00%,其中S14站和S17站均只检出硅藻(图 3)。
枯水期调查结果表明,浮游植物细胞丰度为(4.00~698.40)×104个· L-1,平均为129.21×104个· L-1,也主要集中于万顷沙周围的水道,以S19站最高,其次是S6站(430.13×104个· L-1),S13站最低。细胞丰度组成大部分以硅藻为主,硅藻平均细胞丰度为117.25×104个· L-1,占海域浮游植物平均细胞丰度的90.74%,不同站位硅藻细胞丰度占比为4.88%~100.00%;此外,绿藻在S11站、S13站和S14站都占有较高的比例,裸藻在S17站也同样有较高的比例。
从浮游植物细胞丰度的平均值来看,丰水期的细胞丰度明显高于枯水期。两个时期从各个站位的细胞丰度来看,都是离岸的站位(S6~S9,S18~S20)高于近岸的站位(S1~S5,S10~S13),呈现出近岸向离岸递增的趋势。
2.4 优势种的组成及变化
调查海域浮游植物优势种结果显示,丰水期优势种2种,均为硅藻,新月筒柱藻的优势度最高(0.85);其次是中肋骨条藻,优势度为0.09。枯水期优势种3种,也均为硅藻,其中萎软海链藻(Thalassiosira mala)的优势度最高(0.25);其次是中肋骨条藻(0.20);有棘圆筛藻最低(0.02)。通过对丰水期和枯水期的对比,发现该海域的优势种均为硅藻类,其中中肋骨条藻是两时期的共有优势种,但也表现出季节性的演替现象,丰水期的优势种新月筒柱藻在枯水期更替为萎软海链藻。
2.5 群落多样性
珠江口南沙海域丰水期的H′为0.18~3.32,平均值为1.64;J′为0.07~0.72,平均值为0.40;而该海域枯水期的H′为0.59~2.56,平均值为1.58;J′为0.16~0.64,平均值为0.41(图 4)。丰水期时S4站的H′和J′最高;其次是S5站,分别是2.93和0.68;最低是S17站。枯水期时S1站、S3站、S4站和S10站的H′和J′都较其他站位高;而S9站和S17站则相对较低。
表 3为珠江口南沙海域浮游植物多样性评价结果,表中百分数表示各多样性指标隶属评判阈值的站位数占总站位数的百分比。可见丰水期时种类数、H′和J′都基本处于很低的水平,丰水期的多样性评判结果处于Ⅰ级水平。枯水期时由于H′略高,因此枯水期的多样性评判结果处于Ⅱ级水平。
表 3 珠江口南沙海域浮游植物多样性评价Table 3. Diversity evaluation for phytoplankton in the Nansha sea area of Pearl River Estuary季节
season很低
very low
(Ⅰ)低
low
(Ⅱ)一般
normal
(Ⅲ)高
high
(Ⅳ)很高
very high
(Ⅴ)总评价
general evaluation丰水期wet season 种类数(S)species number 87.50% 12.50% Ⅰ 多样性指数(H′)Shannon-Wiener diversity index 62.50% 37.50% Ⅰ 均匀度指数(J′)Pielous evenness index 100% Ⅰ 枯水期dry season 种类数(S) species number 40% 60% Ⅱ 多样性指数(H′)Shannon-Wiener diversity index 45% 45% 10% Ⅱ 均匀度指数(J′)Pielous evenness index 100% Ⅰ 2.6 浮游植物丰度与环境因子的关系
将浮游植物细胞丰度与水温、盐度进行Pearson相关回归系数分析(表 4),可以看出,丰水期浮游植物细胞丰度与水温呈显著负相关性(P < 0.05);枯水期浮游植物细胞丰度与盐度呈显著正相关性(P < 0.01)。可见温度和盐度对该海域浮游植物的群落结构有较大影响,而其他环境因子与细胞丰度相关性不明显。
表 4 珠江口南沙海域浮游植物细胞丰度与环境因子的相关性Table 4. Correlation between cell density of phytoplankton and environmental factors in the Nansha sea area of Pearl River Estuary季节
season水温
temperature盐度
salinity溶解氧
DO酸碱度
pH丰水期wet season -0.670* -0.209 0.208 0.378 枯水期dry season 0.491 0.597** -0.233 0.133 注:*.P < 0.05;* *.P < 0.01 2.7 典范对应分析
在CCA排序图中(图 5),丰水期与第1排序轴呈正相关的环境因子为盐度(SAL)、温度(TEM)、总氮(DIN)、透明度(SD)和磷酸盐(PO4),相关系数分别为0.978、0.813、0.786、0.567和0.490;与第1排序轴呈负相关的环境因子为溶解氧和pH,相关系数分别为-0.528和-0.018;与第2排序轴呈最大正相关和负相关的环境因子分别为磷酸盐和pH,相关系数分别为0.436和-0.623。枯水期与第1排序轴呈正相关关系的环境因子依次为温度、磷酸盐、盐度、总氮和透明度,相关系数分别为0.846、0.740、0.711、0.702和0.553;与第1排序轴呈负相关的环境因子为pH和溶解氧,相关系数分别为-0.754和-0.439;与第2排序轴呈最大正相关的环境因子为pH,相关系数0.430;透明度与第2排序轴呈最大负相关,相关系数为-0.804。结果表明丰水期和枯水期环境因子对浮游植物群落结构的影响作用存在差异,丰水期环境因子的重要性依次为盐度、温度、总氮、pH、透明度、溶解氧和磷酸盐,枯水期则为温度、透明度、磷酸盐、盐度、总氮、pH和溶解氧。
丰水期浮游植物优势种新月筒柱藻和中肋骨条藻分别位于第三象限和第四象限,新月筒柱藻与温度、总氮和磷酸盐呈负相关,与pH呈正相关;而中肋骨条藻与盐度、温度、总氮、磷酸盐、透明度和pH均呈正相关。枯水期浮游植物优势种萎软海链藻位于第三象限,与pH和透明度呈正相关,与温度和磷酸盐呈负相关;中肋骨条藻位于第四象限,与温度、透明度、磷酸盐、盐度和总氮呈正相关,与pH呈负相关;有棘圆筛藻则位于第一象限,与温度、透明度、pH、磷酸盐、盐度和总氮均呈正相关。
3. 讨论
3.1 浮游植物种类组成
根据珠江口海域浮游植物的特点,可划分为河口类、近岸类、外海类等3大生态类群[8],由于受到珠江水系径流汇入的影响,调查海域的盐度比较低,沿岸带来的营养盐较丰富,因此该海域的浮游植物大量生长和繁殖,种类的组成跨度较大,包括沿岸性种和淡水性种。浮游植物种类组成季节变化明显,丰水期共获得浮游植物37属68种(包括变种和变形),而枯水期共获得浮游植物44属60种(包括变种和变形)。与2008年~2010年在该海域的调查结果相比[10],当前该海域中硅藻类在总种类数中所占比例呈下降趋势,绿藻类所占比例呈现明显上升趋势。
3.2 浮游植物细胞丰度与优势种分布特征
调查发现,该海区浮游植物细胞丰度平面分布在丰水期和枯水期都呈现出由近岸区向离岸区递增的趋势(图 3)。位于龙穴岛东面的S6站、S7站、S8站和S9站以及离岸区的S19站和S20站都是浮游植物平均细胞丰度的高值区,较其他站位平均细胞丰度高,分析发现丰水期离岸高值区站位主要是由硅藻类新月筒柱藻大量繁殖造成的;枯水期离岸高值区站位主要是由硅藻类的萎软海链藻和中肋骨条藻繁殖引起。
优势种在群落中起着主导作用,决定着群落结构中的能量传递和物质循环的途径和方式,支配着群落结构的演替方向[18]。丰水期时的优势种为新月筒柱藻和中肋骨条藻,这2种硅藻都属于广温广盐性藻类,其中新月筒柱藻是一种分布广、高繁殖力的底栖硅藻,也是引发赤潮的一种重要藻类[19],在丰水期其细胞丰度占总细胞丰度的85.15%,特别是S17站,该藻细胞丰度百分比为97.70%;枯水期时的优势种是萎软海链藻、中肋骨条藻和有棘圆筛藻,种类相对较多且优势种发生了更替,由以新月筒柱藻向萎软海链藻和中肋骨条藻优势度并重演替。丰水期时优势种以淡水种和广温广盐种为主,但枯水期时主要是以广温广盐种为主,其中有棘圆筛藻是低温高盐种,这与各个时期由于地表径流引起的水体温度和盐度变化相适应。
丰水期的新月筒柱藻和枯水期的萎软海链藻及中肋骨条藻的细胞丰度分布格局呈现近岸向离岸递增的趋势,这同该海区浮游植物总细胞丰度的分布格局相一致。同时该海域的优势种新月筒柱藻、中肋骨条藻和萎软海链藻都是属于赤潮藻类,说明该海域富营养化严重,有较大的赤潮爆发风险[19]。
3.3 浮游植物群落结构多样性
珠江口南沙海域丰水期和枯水期浮游植物种类数的分布都表现为由河口处向外递减的特征(图 2、图 3),这是由于该海域生长繁殖的主要是以淡水种和广温广盐种为主的藻类[8],盐度的升高反而成为抑制淡水种生长繁殖的因子。藻类平均种类数丰水期略多于枯水期,这是由于丰水期地表径流的增加,导致低盐度的水域扩大,从而使淡水藻类和广温广盐藻类生长繁殖区间扩大,其中丰水期离河口最近的S1站的种类数明显高于同时期的各站位种类数,离河口较远的S19站和S20站的种类数都比较低;而枯水期各站位的种类数较接近(12~16种),这是由于枯水期地表径流的减小,高盐度的海水入侵,导致淡水藻类生长繁殖区间减小,因此以广温广盐类藻类为主。多样性评价结果表明枯水期高于丰水期,但总体而言,两时期的多样性和均匀度都处于较低水平,表明珠江口南沙段海域水质状况一般,水环境受到了一定程度的污染,水域生态系统较为脆弱。
3.4 浮游植物群落结构与水温、盐度的关系
浮游植物群落结构与盐度、水温、营养盐、海流、光照等环境因子存在一定的相关性[20-22],其中水温、盐度是影响浮游植物分布的重要因素[23-25]。广州南沙海域浮游植物群落是以硅藻类为主体的浮游植物群落结构,丰水期和枯水期的细胞丰度都呈现出近岸向离岸递增的趋势。从浮游植物丰度和环境因子的关系,并结合CCA分析可以看出,温度、盐度仍是影响该海区浮游植物的主要环境因子。丰水期细胞丰度与水温、盐度呈负相关性;枯水期细胞丰度则与水温、盐度呈正相关性。由于丰水期浮游植物丰度主要受优势种新月筒柱藻丰度的影响,而新月筒柱藻与温度和盐度呈负相关(图 5-a),这可能是丰水期浮游植物丰度与温度和盐度呈负相关的原因。而枯水期优势种中肋骨条藻和有棘圆筛藻以及浮动弯角藻等多数常见种与温度、盐度呈正相关(图 5-b),而这些种类是枯水期浮游植物丰度的重要组成部分,这可能是浮游植物丰度与温度、盐度呈正相关的主要原因。此外,浮游植物种类不同,其适应的水温和盐度的范围也不同。丰水期时优势种以淡水种和广温广盐种为主,盐度的升高反而抑制了淡水种的生长繁殖,导致细胞丰度的分布格局跟温盐的分布呈现负相关;而枯水期时优势种主要以广温广盐种为主,水温的变化范围为24.7~26.6 ℃,此时细胞丰度与水温呈正相关,表明该水域水温适合大部分藻类的生长,并且随着水温的升高,浮游植物细胞丰度呈增长的趋势,盐度的变化范围为0.14~21.39,适合广温广盐类的藻类生长繁殖,因此枯水期细胞丰度的分布格局与温盐分布格局呈正相关。
4. 小结
1) 调查结果表明,珠江口南沙海域共鉴定浮游植物5门44属76种,种类的组成跨度较大,包括淡水种和广温广盐种,其中硅藻为最主要优势种群。丰水期优势种有2种,分别为新月筒柱藻(0.85)和中肋骨条藻(0.09);枯水期优势种有3种,分别为萎软海链藻(0.25)、中肋骨条藻(0.20)和有棘圆筛藻(0.02)。
2) 丰水期和枯水期浮游植物平均细胞丰度分别为651.91×104个· L-1和129.21×104个· L-1,分别以S17站和S19站最高,而细胞丰度最低的站位位置接近(分别是S14站和S13站)。丰水期和枯水期的细胞丰度分布都表现为近岸向离岸递增的趋势。
3) H′和J′的平均值在丰水期分别为1.64和0.40,而在枯水期分别为1.58和0.41。调查海区的浮游植物群落结构多样性总体处于较低水平,表明珠江口南沙海域水质状况一般,水环境受到了一定程度的污染。
4) 环境因子对珠江口南沙海域浮游植物群落结构有明显影响,丰水期主要影响因子为盐度、温度、总氮和pH;枯水期为温度、透明度、磷酸盐和盐度。
此研究采样频次偏少,今后应加强对其季节演替及动态变化的研究,以更好地理解人类活动干扰下珠江口生态系统的变化过程。
-
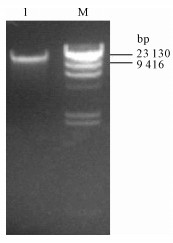
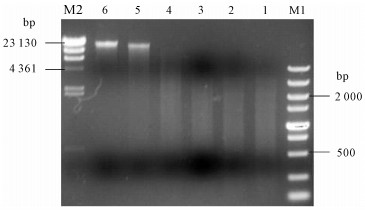
图 2 无乳链球菌基因组DNA酶切Sau3AI酶量的优化
M1. DL5000 DNA Marker;M2. λ/Hind Ⅲ DNA Marker;1~6. 分别使用1 U · μg-1、0.5 U · μg-1、0.25 U · μg-1、0.125 U · μg-1、0.062 5 U · μg-1和0 U · μg-1的Sau3AⅠ酶切基因组DNA
Figure 2. Optimization of volumes of Sau3AⅠ digesting genomic DNA of S.agalactiae
M1. DL5000 DNA Marker; M2. λ/Hind Ⅲ DNA Marker; 1~6. volumes of Sau3AⅠ used to digest genomic DNA were 1 U · μg-1, 0.5 U · μg-1, 0.25 U · μg-1, 0.125 U · μg-1, 0.062 5 U · μg-1and 0 U · μg-1, respectively.
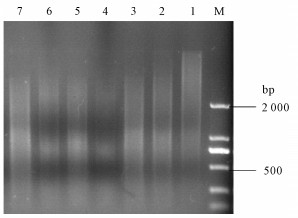
图 3 无乳链球菌基因组DNA酶切时间的优化
M. DL2000 DNA Marker;1~7. Sau3AⅠ酶切基因组DNA的时间分别为5 min、10 min、20 min、30 min、40 min、50 min和60 min
Figure 3. Optimization of time of Sau3AⅠdigesting genomic DNA of S.agalactiae
Optimization of time of Sau3AⅠdigesting genomic DNA of S.agalactiae M. DL2000 DNA Marker; 1~7. Time of genomic DNA digested by Sau3AⅠwere 5 min, 10 min, 20 min, 30 min, 40 min, 50 min and 60 min, respectively.
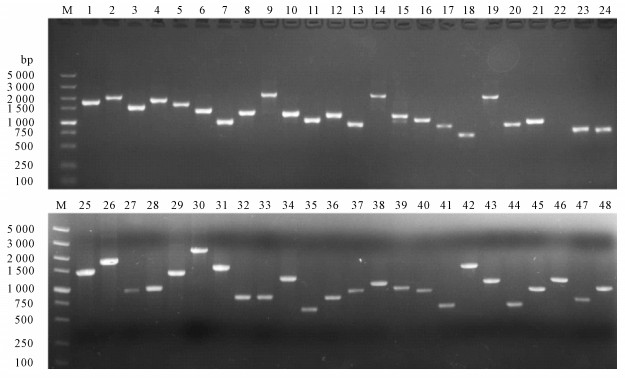
表 1 部分表达文库克隆测序结果与罗非鱼无乳链球菌基因组DNA(GenBank CP003810.1)的同源性
Table 1 Homology between insertion fragments and genomic DNA sequence(GenBank CP003810.1) of S.agalactiae isolated from O.niloticus
克隆编号No. of clone 插入片段大小/bp insertion fragment 同源性/% homology 克隆编号No. of clone 插入片段大小/bp insertion fragment 同源性/% homology 1 1 488 100 16 594 100 2 1 180 100 17 590 100 3 1 089 100 18 575 100 4 1 083 99 19 568 100 5 1 041 100 20 558 100 6 977 99 21 555 99 7 971 100 22 544 100 8 970 99 23 493 100 9 938 100 24 466 100 10 864 100 25 464 99 11 857 100 26 414 100 12 789 100 27 392 100 13 715 100 28 389 100 14 645 100 29 315 100 15 617 100 30 0 0 -
[1] JOHRI A K, PAOLETTI L C, GLASER P, et al. Group B Streptococcus: global incidence and vaccine development[J]. Nat Rev Microbiol, 2006, 4(12): 932-942. doi: 10.1038/nrmicro1552
[2] PERSSON E, BERG S, TROLLFORS B, et al. Serotypes and clinicalm anifestations of invasive group B streptococcal infections in western Seden 1998-2001[J]. Clin Microbiol Infect, 2004, 10(9): 791-796. doi: 10.1111/j.1469-0691.2004.00931.x
[3] PHARES C R, LYNFIELD R, FARLEY M M, et al. Epidemiology of invasive group B streptococcal disease in the United States, 1999-2005[J]. J Am Med Assoc, 2008, 299(17): 2056-2065. doi: 10.1001/jama.299.17.2056
[4] CHAIWARITH R, JULLAKET W, BUNCHOO M, et al. Streptococcus agalactiae in adults at Chiang Mai University Hospital: a retrospective study[J]. BMC Infect Dis, 2011, 11: 149. doi: 10.1186/1471-2334-11-149
[5] DELANNOY C M J, CRUMLISH M, FONTAINE M C, et al. Human Streptococcus agalactiae strain in aquatic mammals and fish[J]. BMC Microbiol, 2013, 13: 41. doi: 10.1186/1471-2180-13-41
[6] 卢迈新, 黎炯, 叶星, 等. 广东与海南养殖罗非鱼无乳链球菌的分离、鉴定与特性分析[J]. 微生物学通报, 2010, 37(5): 766-774. doi: 10.13344/j.microbiol.china.2010.05.022 [7] 祝璟琳, 邹芝英, 李大宇, 等. 尼罗罗非鱼无乳链球菌病的病理学研究[J]. 水产学报, 2014, 38(11): 1937-1944. doi: 10.3724/SP.J.1231.2014.49357 [8] 霍欢欢, 可小丽, 卢迈新, 等. 海南罗非鱼无乳链球菌血清型及耐药性研究[J]. 中国预防兽医学报, 2013, 35(5): 350-354. doi: 10.3969/j.issn.1008-0589.2013.05.03 [9] 祝璟琳, 杨弘, 邹芝英, 等. 海南养殖罗非鱼(Oreochromis niloticus)致病链球菌的分离鉴定及其药敏试验[J]. 海洋与湖沼, 2010, 41(4): 590-596. https://d.wanfangdata.com.cn/periodical/Ch9QZXJpb2RpY2FsQ0hJTmV3UzIwMjQxMTA1MTcxMzA0Eg5oeXloejIwMTAwNDAxORoINDhiZGoydGk%3D [10] 沈定树, 周雪艳. 无乳链球菌表面蛋白的研究进展[J]. 中国微生态学杂志, 2007, 19(5): 472-473. doi: 10.3969/j.issn.1005-376X.2007.05.034 [11] 汪开毓, 付希, 肖丹, 等. 罗非鱼源无乳链球菌cpsE基因的克隆和分子特性分析[J]. 水产学报, 2011, 35(5): 660-665. doi: 10.3724/SP.J.1231.2011.17332 [12] 黎炯, 叶星, 可小丽, 等. 罗非鱼无乳链球菌Sip基因的克隆、表达及免疫原性分析[J]. 水生生物学报, 2012, 36(4): 626-633. doi: 10.3724/SP.J.1035.2012.00626 [13] 李庆勇, 可小丽, 卢迈新, 等. 罗非鱼无乳链球菌C5a肽酶(Scpb)的原核表达及其免疫原性[J]. 中国水产科学, 2014, 21(1): 169-179. doi: 10.3724/SP.J.1118.2014.00169 [14] 王蓓. 基于全基因组序列分析筛选罗非鱼源无乳链球菌ZQ0910株保护性抗原及其免疫保护性研究[D]. 青岛: 中国海洋大学, 2013: 5-8. 10.7666/d.D328905 [15] HANDFIELD M, BRADY L J, PROGULSKE-FOX A, et al. IVIAT: a novel method to identify microbial genes expressed specifically during human infection[J]. Trends Microbiol, 2000, 8(7): 336-339. doi: 10.1016/S0966-842X(00)01775-3
[16] GU H W, ZHU H D, LU C P. Use of in vivo-induced antigen technology (IVIAT) for the identification of Streptococcus suis serotype 2 in vivo-induced bacterial protein antigens[J]. BMC Microbiol, 2009, 9: 201. doi: 10.1186/1471-2180-9-201
[17] 李霆, 高志勇, 王恒, 等. 应用体内诱导抗原技术筛选结核分枝杆菌体内表达基因[J]. 遗传学报, 2005, 32(2): 111-117. https://kns.cnki.net/kcms2/article/abstract?v=8iF5fUaBjd5JvEOhufDhA66gcHnnzutcV7fLbTHM-kLJXz4poQHcY15z8-fvK3TklUACzCSW5XjTecgmeojs_wCnH0yjcN_CpaqLKjPp2PIV6HgmVcPK4uIlSIfz5ENv6huI7EqtxM20N_hB0QcheWmsLazJYm6B2KhCaoyzkXMlsNcQeBfgog-uvvwAl_0v&uniplatform=NZKPT&language=CHS [18] 胡勇, 丛延广, 黎庶, 等. 伤寒沙门菌感染过程中体内诱导表达蛋白抗原的筛选[J]. 中国科学, 2009, 39(9): 879-884. https://kns.cnki.net/kcms2/article/abstract?v=8iF5fUaBjd4SqklRIyURaAEv9jFG7moaUwO6qeBRzHP0L-fVNZAV2y1qaLZkJOEsH7AjD29WVDsDOj4dGh4Qw9LanfLCJsuM-YjR2At1opBBDDftF9sHkZmXVkiHVgnCj2HlOPUdK45fFtC73Fj3T2graxz8wjU_46REFgqXbqLxrjmItKjEVqYQSQ4dwU8M&uniplatform=NZKPT&language=CHS [19] 都启晶. 副猪嗜血杆菌基因组表达文库的构建与其荚膜多糖输出蛋白基因的筛选和免疫原性研究[D]. 雅安: 四川农业大学, 2010. https://kns.cnki.net/kcms2/article/abstract?v=8iF5fUaBjd7d1Si5uKDjEryK_b29iiDlipF8PF8l4tKakFbN5P9HcBCpCrwaTlQ_NXNGekX8PaUj0hij1iYBg7g6YwdSU9yDbAC2EQGOrOPahx2ZPF4YZUY_Auj97udsfnIt-waCwuZkmKxlhs2s22TgIzqpR2bSCrnlWjbGfz2DoinJ6Bv7-mQEGRKrJEMU&uniplatform=NZKPT&language=CHS [20] LI Q C, HU Y C, CHEN J, et al. Identification of Salmonella enterica aerovar pullorum antigenic determinants expressed in vivo[J]. Infect Immun, 2013, 81(9): 3119-3127. doi: 10.1016/j.vaccine.2010.03.071
[21] ROLLINS S M, PEPPERCORN A, HANG L, et al. In vivo induced antigen technology (IVIAT)[J]. Cell Microbiol, 2005, 7(1): 1-9. doi: 10.1111/j.1462-5822.2004.00477.x
[22] SUN Y, HUA Y H, LIU C S, et al. Construction and analysis of an experimental Streptococcus iniae DNA vaccine[J]. Vaccine, 2010, 28(23): 3905-3912. doi: 10.1016/j.vaccine.2010.03.071
[23] 胡元庆, 黄金林, 李求春, 等. 空肠弯曲菌NCTC11168基因组表达文库的构建和鉴定[J]. 中国兽医科学, 2011, 41(11): 1106-1110. doi: 10.16656/j.issn.1673-4696.2011.11.002 -
期刊类型引用(9)
1. 张良奎,林雅君,向晨晖,黄亚东,郑传阳,宋星宇. 粤港澳大湾区近海浮游植物群落结构特征及其驱动因素. 生态科学. 2024(04): 1-10 .  百度学术
百度学术
2. 高广银,李学恒,唐灵刚,李颖颖. 秋冬季浮游植物群落结构特征及其环境因子关系——以珠江口南沙海域为例. 绿色科技. 2024(20): 74-78+84 .  百度学术
百度学术
3. 陈亮东,詹建坡,王庆. 珠江典型河段浮游植物群落结构特征及其对水质的指示作用. 南方水产科学. 2023(06): 1-10 .  本站查看
本站查看
4. 蒋佩文,李敏,张帅,陈作志,徐姗楠. 基于环境DNA宏条码和底拖网的珠江河口鱼类多样性. 水生生物学报. 2022(11): 1701-1711 .  百度学术
百度学术
5. 李桂娇,冯洁娉,叶锦韶. 南沙港海区水质变化与浮游植物群落更替特征. 生态科学. 2021(05): 164-171 .  百度学术
百度学术
6. 姚艳欣,陈楠生. 珠江口及其邻近海域赤潮物种的生物多样性研究进展. 海洋科学. 2021(09): 75-90 .  百度学术
百度学术
7. 陈思,李艺彤,蔡文贵,陈海刚,田斐,张林宝,张喆,郭志勋. 虾蟹混养池塘浮游植物群落结构的变化特征. 南方水产科学. 2020(03): 79-85 .  本站查看
本站查看
8. 李亚军,王先明,程贤松,魏盟智,李江月,朱为菊,邓晓东. 海南岛海尾湾浮游植物群落结构及水质的调查. 热带生物学报. 2020(03): 257-265 .  百度学术
百度学术
9. 李希磊,于潇,卢钰博,杨俊丽,崔龙波. 2015—2017年莱州湾招远海域扇贝养殖区浮游藻类群落变化. 南方水产科学. 2019(04): 11-18 .  本站查看
本站查看
其他类型引用(4)




 下载:
下载:




 粤公网安备 44010502001741号
粤公网安备 44010502001741号
