Species composition and succession of coral reef fish in Yuzhuo Reef, Xisha Islands
-
摘要:
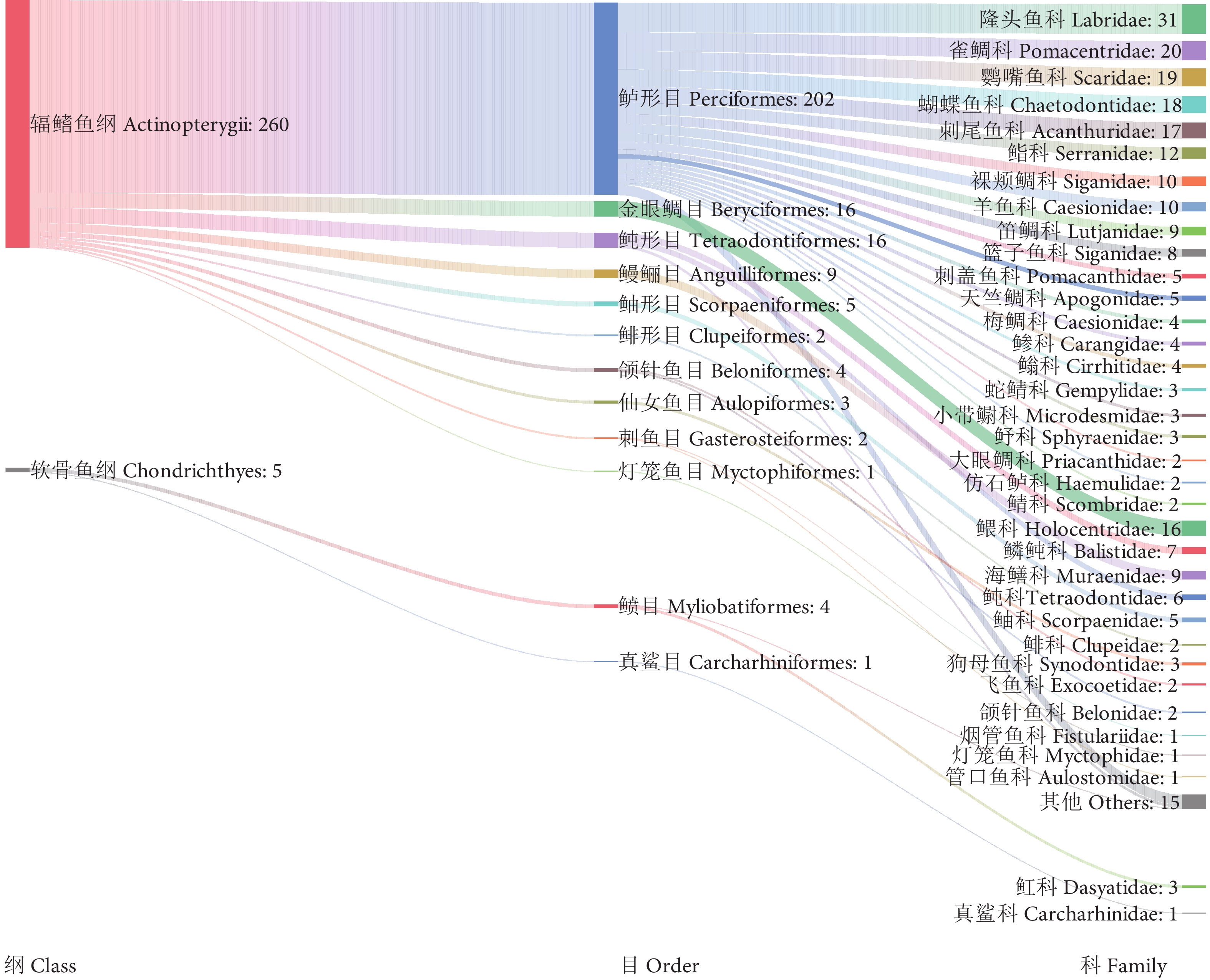
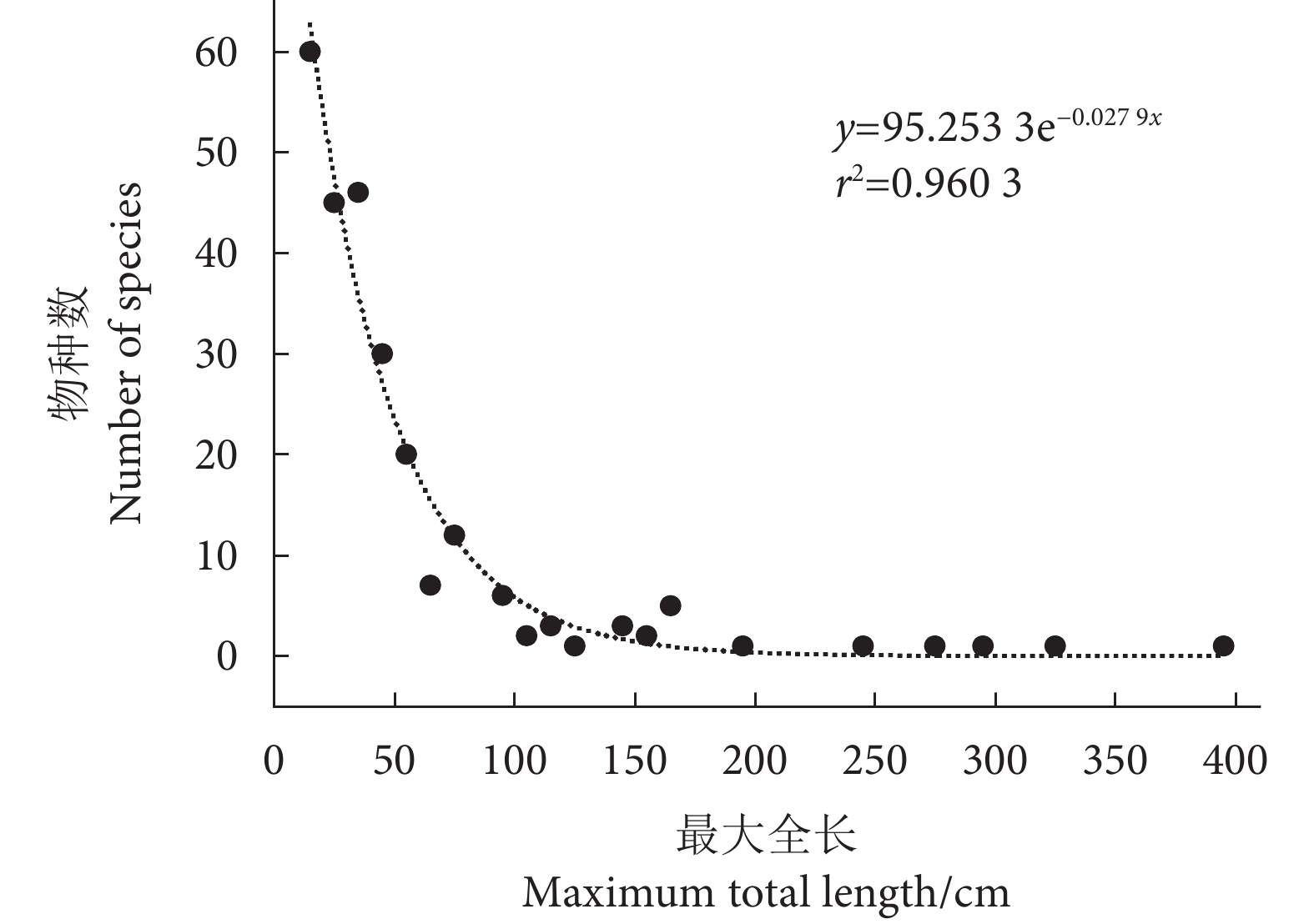
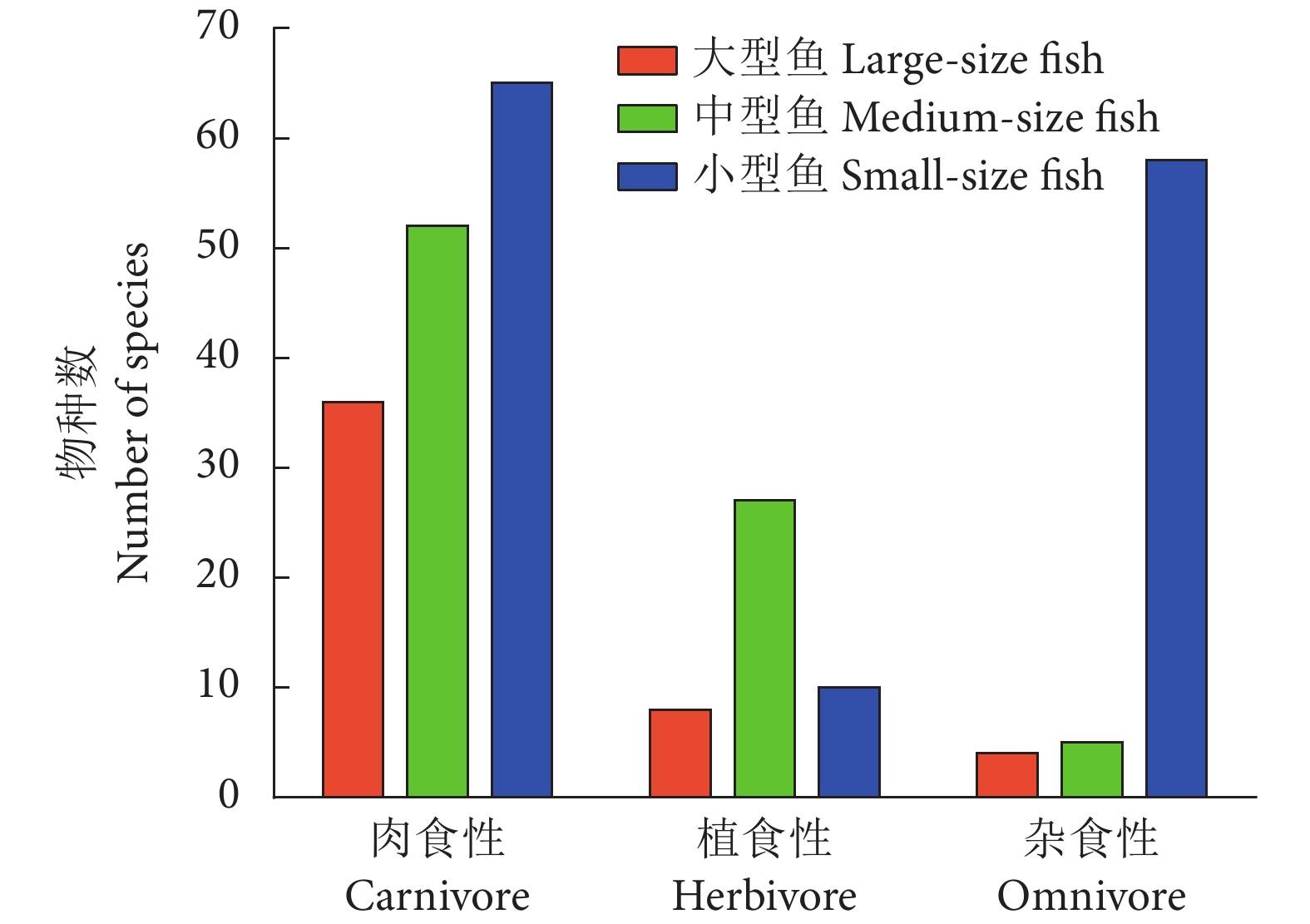
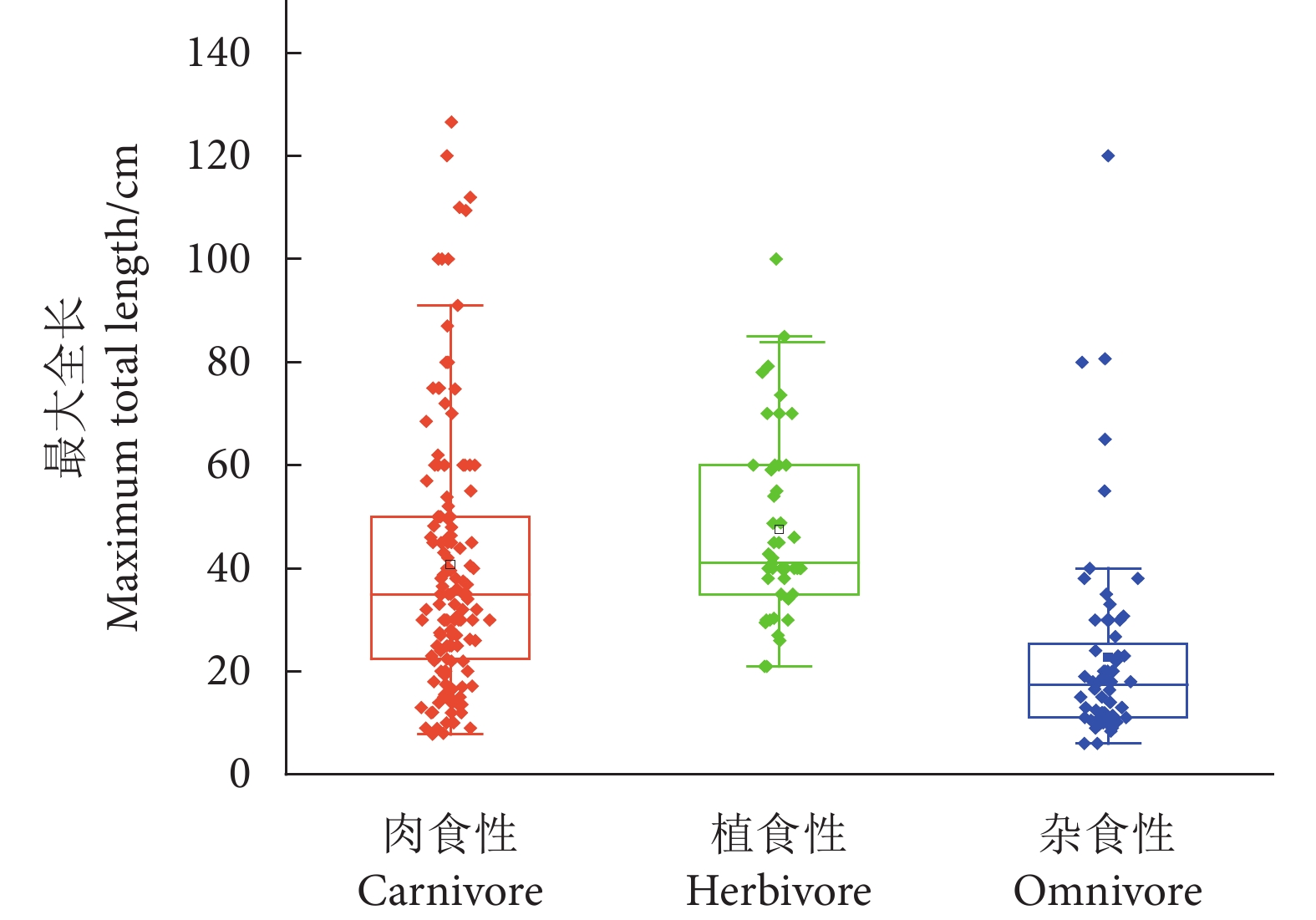
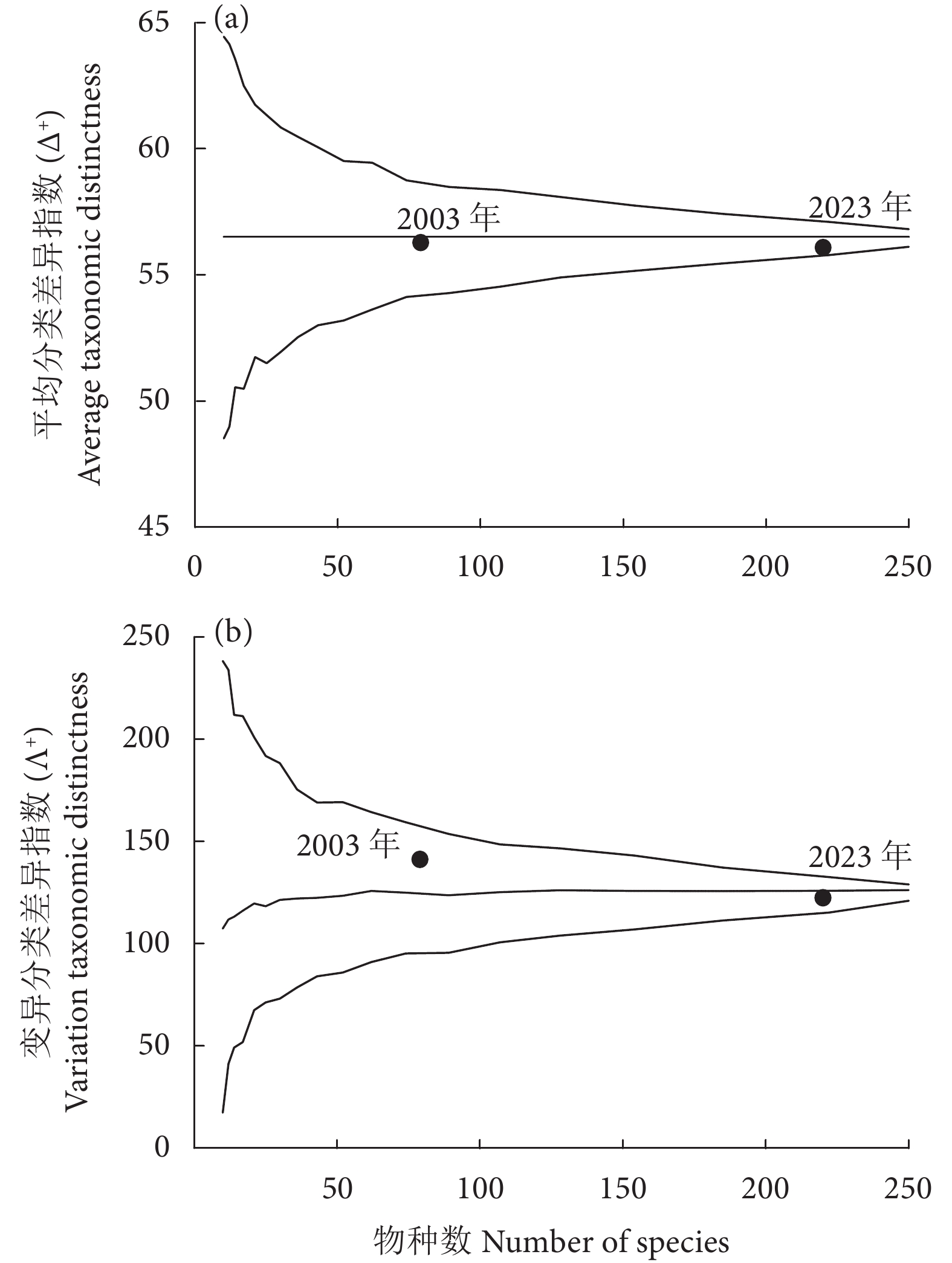
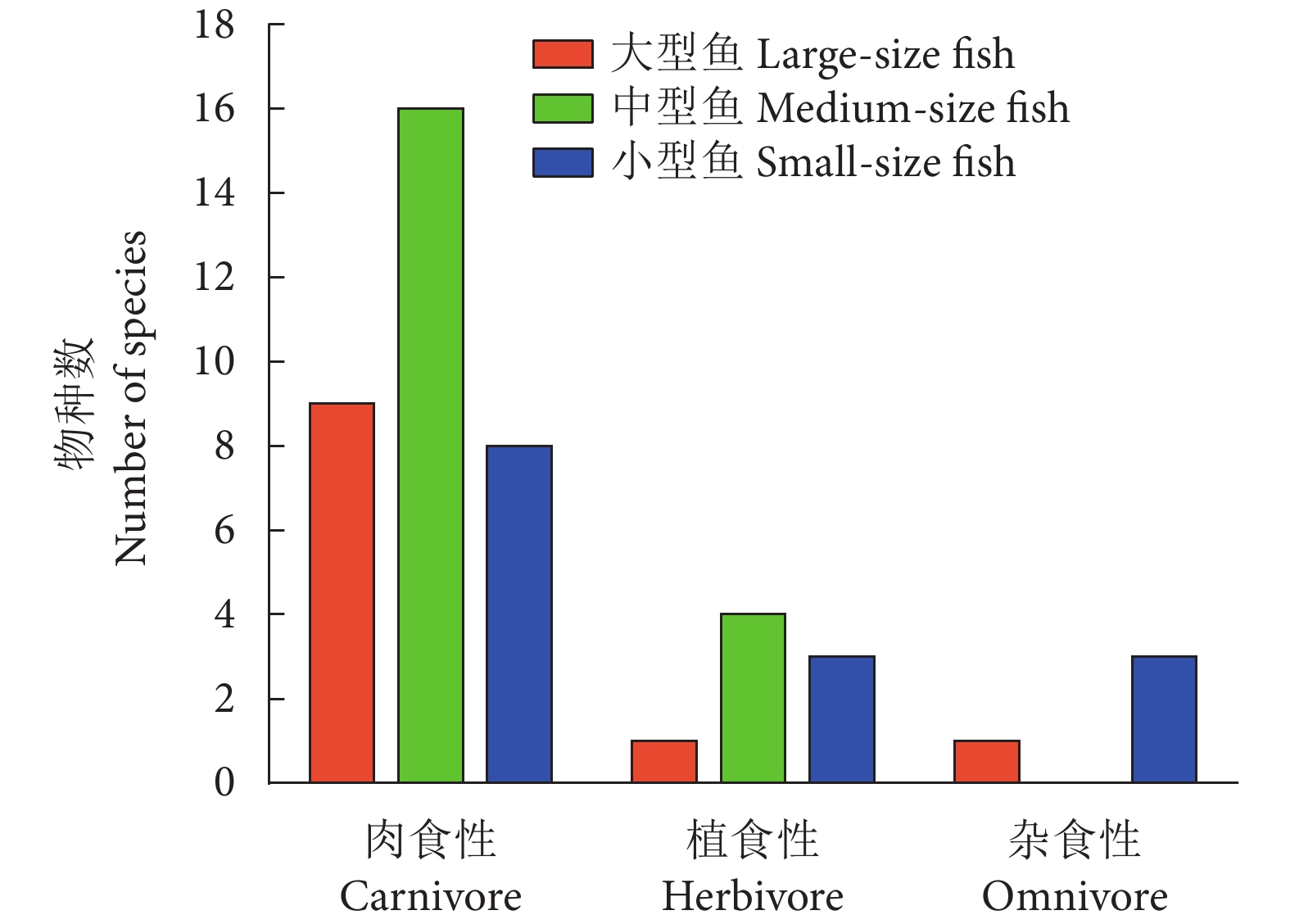
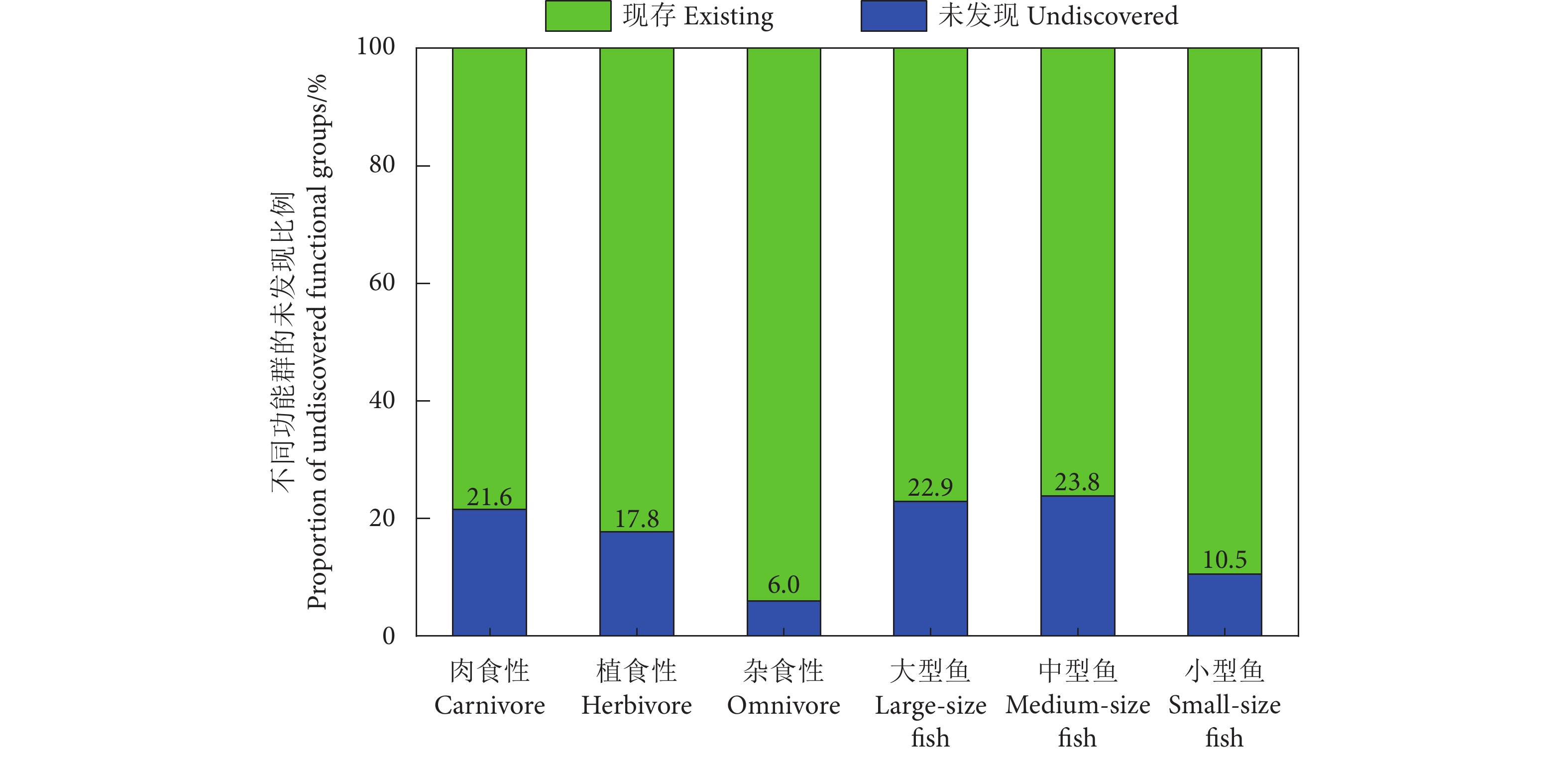
玉琢礁是西沙群岛中的重要环礁,拥有多样化的珊瑚礁生态系统和丰富的鱼类资源。为掌握玉琢礁珊瑚礁鱼类的资源状况及其演变特征,于2023年采用手钓、水下潜捕、水下视频和eDNA等方法,对其珊瑚礁鱼类资源进行调查。共发现鱼类220种,其中eDNA方法共发现鱼类111种,结合2003年中国水产科学研究院南海水产研究所的历史档案数据,截至目前在玉琢礁共发现鱼类265种,隶属于12目50科128属,其中鲈形目占绝对优势。鱼类群落组成以小型鱼类为主,平均分类差异指数 (Δ+) 和变异分类差异指数 (Λ+) 分别为56.08和122.4;相似性分析表明,2003与2023年之间,鱼类在种类组成、食性组成和不同体型层面的相似性均表现为不相似或极不相似。与历史调查数据进行比较,共有45种鱼类未被监测到,其中大型肉食性鱼类受到自然与人类扰动的影响最为显著,植食性鱼类所受到的影响相对较小,分类多样性指数 (Δ+、Λ+) 显著下降。初步推断不可持续的捕捞、生境衰退和温度上升等因素导致了玉琢礁珊瑚礁鱼类的更替。研究结果可为玉琢礁的珊瑚礁鱼类资源保护与管理、珊瑚礁生态系统的恢复提供参考。
Abstract:Yuzhuo Reef is an important atoll in the Xisha Islands, with diverse coral reef ecosystems and rich fish resources. In order to understand the status of coral reef fish resources in Yuzhuo Reef and their evolutionary features, we investigated the fish resources of Yuzhuo Reef in 2023 through methods such as handline fishing, underwater spearing, underwater videography and environmental DNA (eDNA) analysis, identifying 220 fish species in total, of which 111 were discovered through the eDNA method alone. Combining with the historical archival data from the South China Sea Fisheries Research Institute in 2003, we had already recorded a total of 265 fish species in Yuzhuo Reef, which belong to 12 orders, 50 families and 128 genera, with Perciformes species dominating the species composition. The fish community composition was primarily characterized by small-size fish, with the average taxonomic distinctness index (Δ+) and variation in taxonomic distinctness index (Λ+) being 56.08 and 122.4, respectively. Similarity analyses show that from 2003 to 2023, the similarity in species composition, dietary habits and body size was either dissimilar or extremely dissimilar. Compared with the historical records, 45 fish species were not observed; particularly large carnivorous fish were most significantly affected by natural and human disturbances. Conversely, the impact on phytophagous fish was relatively small, with siginificant decline in taxonomic diversity indexes (Δ+, Λ+). It is suggested that unsustainable fishing practices, habitat degradation and rising temperatures have contributed to changes in the fish community structure in Yuzhuo Reef. The findings provide references for the conservation and management of coral reef fish resources and the rehabilitation of coral reef ecosystems in Yuzhuo Reef.
-
Keywords:
- Xisha Islands /
- Coral reef fish /
- Human disturbance /
- Habitat decline /
- Evolution
-
紫菜(Porphyra)属于红藻门、红毛菜科,已经报道有134个物种,广泛分布于从寒带到亚热带的潮间带海域。紫菜是一类重要的经济红藻,作为目前世界上人工养殖海藻中经济价值最高的种类,仅中、日、韩三国紫菜的初级加工品年产值就超过20×109美元[1]。中国是世界紫菜的主要生产国和出口国,主要栽培种类包括条斑紫菜(P.yezoensis)和坛紫菜(P.haitanensis)。长期以来紫菜的遗传育种研究以诱变育种、单性生殖、选择育种等经典育种技术为主,相对于蓬勃发展的动植物分子育种研究而言,分子标记技术尤其是共显性分子标记数量少、应用种类有限,制约了紫菜遗传改良工作的快速发展。微卫星又称为简单序列重复(simple sequence repeat,SSR),作为一种共显性遗传标记,具有多态性高、重复性好等优点[2],已被广泛应用到海带属(Laminaria; Saccharina)[3-4]、江蓠属(Gracilaria)[5]、龙须菜属(Gracilariopsis)[6]、紫菜属(Porphyra)[7-8]等重要大型海藻的遗传多样性、遗传图谱构建和QTL定位研究中。
皱紫菜(P.crispata)是中国南方沿海野生紫菜的主要种类之一,具有重要的经济价值和药用价值。研究表明,皱紫菜的高温耐受性较坛紫菜和条斑紫菜要强,对环境具有更强的适应能力[9]。目前中国已经进行皱紫菜人工栽培试验,增加紫菜栽培新种类对于解决坛紫菜栽培生产中因高温而出现“烂菜”等问题具有重要的意义。半叶紫菜(P.katadai)和少精紫菜(P.oligospermatangia)均为野生种类,主要分布于北方沿海地区,和条斑紫菜都属于雌雄同体的紫菜种类,也是紫菜属遗传学研究的良好试验材料。微卫星标记在紫菜属不同物种中的研究应用较少,关于皱紫菜、半叶紫菜和少精紫菜等紫菜属物种的微卫星标记尚未见报道。笔者利用微卫星DNA分子标记技术分析皱紫菜等5个紫菜属物种的遗传结构及种质,以便更有效地推动紫菜种质资源评价并辅助紫菜遗传改良,同时对微卫星标记在紫菜属物种遗传分析中的通用性进行探讨,以期为紫菜属的分子遗传学研究提供依据。
1. 材料与方法
1.1 试验材料
试验所用的紫菜属5个物种10个品系自由丝状体(表 1)由中国海洋大学汤晓荣老师惠赠,培养于中国海洋大学海洋生物遗传育种研究室。
表 1 此试验所用的10个品系紫菜丝状体信息表Table 1 Information of 10 lines of Porphyra conchocelis in this study序号
No.品系代码
line code物种名称
species name1 PH1 坛紫菜P.haitanensis 2 PH2 坛紫菜P.haitanensis 3 PH3 坛紫菜P.haitanensis 4 PH4 坛紫菜P.haitanensis 5 PK1 半叶紫菜P.katadai 6 PK2 半叶紫菜P.katadai 7 PY1 条斑紫菜P.yezoensis 8 PY2 条斑紫菜P.yezoensis 9 PO 少精紫菜P.oligospermatangia 10 PC 皱紫菜P.crispata 1.2 基因组DNA的提取
紫菜丝状体基因组DNA的提取均采用植物基因组DNA提取试剂盒(北京天根)法提取,用Nanodrop 1000分光光度计测定DNA的光密度(OD)和浓度,并在1.0%琼脂糖凝胶上电泳(100 V 30 min)检测,用JS-380A自动凝胶图像分析仪拍照并进一步检验DNA的质量和浓度。
1.3 微卫星引物合成及筛选
现有的文献报道紫菜微卫星引物共68对:25对引自SUN等[10]发布的微卫星引物,10对引自刘必谦等[11]发布的微卫星引物,11对引自ZUO[12]发布的微卫星引物,8对引自XIE等[13]发布的微卫星引物,14对引自KONG等[14]发布的微卫星引物,引物由华大基因公司合成。以5个不同物种的丝状体(PH2, PK2, PY2, PO和PC)DNA样品为模板,对所有引物进行PCR筛选,从中选择扩增稳定且条带清晰的微卫星引物(引物信息见表 2)。
表 2 20对微卫星引物序列和特异退火温度Table 2 Sequence of 20 pairs of microsatellite marker primers and specific annealing temperature for PCR amplification引物编号
primer No.引物序列(5′→3′)
primer sequence重复序列
repetitive sequence退火温度/℃
annealing temperature片段范围
fragment rangeGenBank登录号
GenBank accession No.参考文献
referenceSSR-ZC1 TTCGCTGCGTTTCACCTTACATTT (TGCG)6 60 205 AV434771 [10] ACAAGGCCAACCCGAACACA SSR-ZC2 GGCTGCGGCTGAGTCACAGA (AGC)8 60 235 AU194221 [10] GTCGCTCCAACTCCTCCTGCT SSR-ZC3 TGGTGCTGTCTTCCAACGAGTA (AGC)13 60 245 AU196122 [10] CGGCTGTCGCACCTCGTTATA SSR-ZC7 CGCTCAACCACTTCGTCAG (AAC)10+1 58 261 AU191538 [10] CATTGTTGGCGTTGTTGTCATA SSR-ZC9 CCGTCGTCAGCAGGAGCA (GGC)7 60 200 AV434560 [10] ATGTGAGAAGCCAGTAGGGAAAGT SSR-ZC10 ACTTCCATCGCTGTCTTCGCT (AGC)8 60 196 AU192094 [10] CTGAGCTGCGTGTTGTGGTT SSR-ZC12 TCAACCATCAGCCATACCGAC (ACC)11 58 192 AV435510 [10] GACATGTCCGCCACCTTGTA SSR-ZC15 TGACTTCCTCATCGACATTGT (AGC)8 55 319 AU187439 [10] GCCATAGTACATTTGTTGCTG SSR-ZC16 TACCAGGTCGACCAGGAGCA (AGC)11 60 291 AU193458 [10] TCCAACTCTGCAGTGTCCGTT SSR-ZC19 ACCTCCTCGGCTACTTCAGA (AGC)7 58 276 AV431181 [10] GGATACAACGCCTGCTCCAT SSR-ZC20 GGCAGCAGCCATGATGTA (AGC)8+8 55 339 AV433675 [10] CCGTCAGGCAGAAGAGAAT SSR-ZC23 CAAGGGCTACTGCTACTA CAA (AAC)17 55 249 AU195299 [10] TACAAAAAGACTCTCGTG GCA SSR-ZC25 CCGTGCTACTACGGCTA CAA (AAC)9+11 58 302 DN606134 [10] GTCCGGTGCAGGTTG TTCT SSR-ZC36 CCAACGATGGGTTTCTTCAA 55 300 [7] ACTTCATGCCCCTGCCGATG SSR-ZC38 GGACAAGGGGTAATGGCT (CA)17 50 212~226 DQ831155 [12] TGGAAAACTTCCTGGGTG SSR-ZC43 GGAATGCCTTGCACCTGG (GT)26 55 408~420 DQ831185 [12] CACTATTGACCGAATCCGCTAC SSR-ZC44 CGTGCGAGTCATAGTCTGCT (GTGA)26 55 191~200 DQ831194 [12] ACAGCCAGTGCAAGAACACC SSR-ZC47 CTAGACGGAGTGCGGCTGAC (CA)9 58 176~196 DQ831197 [12] GCTCCTCCACGAGCATCAGG SSR-ZC54 GGTGGCAGTGAAGCGAAACA (CG)7 60 229 DN607742 [13] ACCCAGCAAGTGCGTGAGC SSR-ZC61 AACAGAGATACGGAGAGC (GT)8..(TG)11 50 276 EU670707 [14] ATCAGATTGGACTTGCCT 1.4 PCR扩增及电泳
利用筛选出的微卫星引物对5个物种的10个丝状体品系进行PCR扩增。
PCR反应在PCR仪(BIOER)上进行。反应体系为20 μL,试剂终浓度分别为1.5 mmol·L-1,10×PCR buffer[含氯化镁(MgCl2)], 0.125 mmol·L-1dNTP, 20 ng模板DNA,0.05 U TaqDNA聚合酶,微卫星引物1.0 μL。
PCR反应程序为94 ℃变性5 min;接35个循环为94 ℃ 50 s,退火45 s(退火温度因引物而定),72 ℃ 1 min;72 ℃最后延伸10 min, 4 ℃保存,1.0%琼脂糖电泳,100 V 50 min,检测PCR产物。取部分选扩产物(3 μL)与等体积的上样缓冲液(3 μL)充分混匀,95 ℃变性5 min。点样6 μL,在6%变性聚丙烯酰胺凝胶上电泳,仪器为DYY-12C电泳仪(北京市六一仪器厂出品),电泳前进行30 min预电泳,最大功率恒功率电泳2 h。银染程序参照SANGUINETTI等[15]报道的方法。室温下自然干燥,用扫描仪扫描并保存图像。
1.5 数据统计分析
微卫星标记按共显性标记进行数据统计及分析。根据分子量大小对扩增结果读带,以二倍体形式记录,从大到小依次记为A,B,C……。利用POPGENE32(V 1.31) (ftp://ftp.microsoft.com/Softlib/MSLFILES/HPGL.EXE)软件计算各群体的观测有效等位基因数(Ne)、多态位点百分率(P)、基因多样性指数(Nei′s)、Shannon信息指数(I)、表观杂合度(Ho)和预期杂合度(He)、Nei′s标准遗传距离(D)、遗传相似度和遗传分化指数(Fst)。根据Nei′s标准遗传距离,利用MEGA 4.0 (http://www.megasoftware. net/)中的非加权平均算术法(UPGMA)对各种群进行聚类分析,自展检验1 000次。
2. 结果与分析
2.1 紫菜丝状体基因组DNA的提取
所提取的紫菜丝状体基因组DNA,OD260/OD280为1.7~1.9,质量浓度为20~40 ng·μL-1,没有DNA降解、RNA残留及蛋白质污染,可以满足微卫星遗传分析的需要(图 1)。
![]() 图 1 紫菜丝状体基因组DNA在1.0%的琼脂糖电泳检测结果M.DL 2 000分子量标准;1~4. PH1、PH2、PH3、PH4,坛紫菜; 5~6. PK1、PK2,半叶紫菜; 7~8. PY1、PY2, 条斑紫菜;9. PO,少精紫菜;10.PC, 皱紫菜Fig. 1 Electrophoresis in 1% agarosegel of genomic DNA obtained from conchocelis of PorphyraM. DL 2 000 DNA marker; 1~4. PH1, PH2, PH3, PH4, P.haitanensis; 5~6. PK1, PK2, P.katadai; 7~8. PY1, PY2, P.yezoensis; 9. PO, P.oligospermatangia; 10. PC, P.crispata
图 1 紫菜丝状体基因组DNA在1.0%的琼脂糖电泳检测结果M.DL 2 000分子量标准;1~4. PH1、PH2、PH3、PH4,坛紫菜; 5~6. PK1、PK2,半叶紫菜; 7~8. PY1、PY2, 条斑紫菜;9. PO,少精紫菜;10.PC, 皱紫菜Fig. 1 Electrophoresis in 1% agarosegel of genomic DNA obtained from conchocelis of PorphyraM. DL 2 000 DNA marker; 1~4. PH1, PH2, PH3, PH4, P.haitanensis; 5~6. PK1, PK2, P.katadai; 7~8. PY1, PY2, P.yezoensis; 9. PO, P.oligospermatangia; 10. PC, P.crispata2.2 引物筛选结果
从68对微卫星引物中筛选出20对扩增稳定、多态性高的引物(表 2),其部分扩增结果见图 2。其中有14对引物在5个物种中均可扩增出稳定的条带,扩增片段大小为192~420 bp,且这14对引物中有10对可在10个丝状体中扩增出稳定的条带。
![]() 图 2 部分微卫星引物(左:SSR-ZC15;右:SSR-ZC20)在10个紫菜丝状体中的扩增结果M.DL100分子量标准;1~10. PH1,PH2,PH3,PH4,PK1,PK2,PY1,PY2,PO,PCFig. 2 Electrophoresis of part of microsatellite primer-pairs (L: SSR-ZC15; R: SSR-ZC20) in 10 lines of Porphyra conchocelisM. DNA maker DL100; 1~10. PH1, PH2, PH3, PH4, PK1, PK2, PY1, PY2, PO, PC
图 2 部分微卫星引物(左:SSR-ZC15;右:SSR-ZC20)在10个紫菜丝状体中的扩增结果M.DL100分子量标准;1~10. PH1,PH2,PH3,PH4,PK1,PK2,PY1,PY2,PO,PCFig. 2 Electrophoresis of part of microsatellite primer-pairs (L: SSR-ZC15; R: SSR-ZC20) in 10 lines of Porphyra conchocelisM. DNA maker DL100; 1~10. PH1, PH2, PH3, PH4, PK1, PK2, PY1, PY2, PO, PC2.3 有效等位基因数和遗传杂合度
10个紫菜丝状体个体间的平均Ne为2.37,平均He为0.550 9,I为0.899 9,Fst为0.400 0~1.000 0,平均为0.733 3。10个品系的紫菜丝状体具有良好的遗传多样性,且不同材料之间遗传分化较明显(表 3)。
表 3 20个微卫星的遗传多样性指数Table 3 Genetic diversity index of 20 microsatellite locilocus 表观杂合度
Ho预期杂合度
He有效等位基因数
NeShannon信息指数
I遗传分化指数
Fst基因流
NmSSR-ZC1 0.700 0 0.647 4 2.60 1.010 4 0.430 9 0.330 2 SSR-ZC2 1.000 0 0.733 3 2.57 1.011 4 0.844 6 0.046 0 SSR-ZC3 0.333 3 0.333 3 1.38 0.450 6 0.946 5 0.014 1 SSR-ZC7 0.000 0 0.484 8 1.80 0.636 5 1.000 0 0 SSR-ZC9 0.000 0 0.189 5 1.22 0.325 1 1.000 0 0 SSR-ZC10 0.100 0 0.721 1 3.17 1.565 4 0.927 0 0.019 7 SSR-ZC12 0.600 0 0.526 3 2.00 0.693 1 0.400 0 0.375 0 SSR-ZC15 0.100 0 0.415 8 1.65 0.687 4 0.873 4 0.036 2 SSR-ZC16 0.444 4 0.679 7 2.79 1.161 6 0.718 3 0.098 0 SSR-ZC19 0.666 7 0.797 4 4.05 1.565 4 0.625 0 0.150 0 SSR-ZC20 0.784 2 0.784 2 3.92 1.449 0 0.932 9 0.018 0 SSR-ZC23 0.500 0 0.575 0 2.17 0.921 5 0.716 3 0.099 0 SSR-ZC25 0.600 0 0.563 2 2.15 0.845 1 0.439 3 0.319 1 SSR-ZC36 0.111 1 0.385 6 1.57 0.654 7 0.896 9 0.028 7 SSR-ZC38 0.500 0 0.763 2 3.64 1.415 0 0.655 2 0.131 6 SSR-ZC43 0.666 7 0.732 0 3.24 1.239 5 0.600 0 0.166 7 SSR-ZC44 0.571 4 0.571 4 2.13 0.991 1 0.740 3 0.087 7 SSR-ZC47 0.285 7 0.439 6 1.69 0.598 3 0.859 2 0.041 0 SSR-ZC54 0.100 0 0.542 1 2.06 0.823 7 0.902 9 0.026 9 SSR-ZC61 0.500 0 0.684 2 2.86 1.161 1 0.615 4 0.156 3 平均mean 0.375 2 0.550 9 2.37 0.899 9 0.733 0 0.091 1 标准差
standard deviation0.288 5 0.205 5 0.872 7 0.392 6 2.4 遗传距离、遗传相似指数和聚类分析
10个品系紫菜丝状体的遗传相似指数为0.254 6~0.899 4,平均为0.474 9,最小值出现在皱紫菜(PC)和半叶紫菜(PK1)中,最大值出现在条斑紫菜的2个不同品系中;遗传距离为0.106 0~1.406 6,平均为0.792 3,最大值出现在坛紫菜(PH1)和少精紫菜(PO)中,最小值出现在条斑紫菜的2个不同品系中(表 4)。
表 4 10个紫菜品系遗传距离(左下方)和相似性系数(右上方)矩阵Table 4 Genetic distance (lower-trangular data matrix) and genetic similarity (upper-triangular data matrix) of 10 Porphyra linesPOP ID 坛紫菜1
PH1坛紫菜2
PH2坛紫菜3
PH3坛紫菜4
PH4半叶紫菜1
PK1半叶紫菜2
PK2条斑紫菜1
PY1条斑紫菜2
PY2少精紫菜
PO皱紫菜
PC坛紫菜1 PH1
P.haitanensis 10.445 4 0.471 9 0.506 0 0.307 7 0.302 2 0.296 9 0.373 1 0.245 0 0.423 1 坛紫菜2 PH2
P.haitanensis 20.808 9 0.753 8 0.619 8 0.335 0 0.296 1 0.348 5 0.377 2 0.384 7 0.477 0 坛紫菜3 PH3
P.haitanensis 30.751 1 0.282 7 0.860 8 0.555 6 0.527 4 0.485 8 0.465 1 0.391 2 0.436 4 坛紫菜4 PH4
P.haitanensis 40.681 2 0.478 4 0.149 8 0.474 3 0.414 0 0.444 9 0.380 4 0.338 9 0.379 5 半叶紫菜1 PK1
P.katadai 11.178 5 1.093 6 0.587 8 0.745 8 0.872 9 0.502 5 0.465 1 0.425 3 0.254 6 半叶紫菜2 PK2
P.katadai 21.196 7 1.217 1 0.639 9 0.881 8 0.136 0 0.526 4 0.488 2 0.467 7 0.357 1 条斑紫菜1 PY1
P.yezoensis 11.214 3 1.054 2 0.722 0 0.809 8 0.688 1 0.641 8 0.899 4 0.692 4 0.493 5 条斑紫菜2 PY2
P.yezoensis 20.985 9 0.975 1 0.765 5 0.966 6 0.765 5 0.717 0 0.106 0 0.721 8 0.519 7 少精紫菜PO
P.oligospermatangia1.406 6 0.955 4 0.938 4 1.082 1 0.855 1 0.759 9 0.367 6 0.326 0 0.567 9 皱紫菜PC
P.crispata0.860 2 0.740 2 0.829 1 0.968 8 1.368 1 1.029 6 0.706 3 0.654 5 0.565 8 根据各个体间的遗传距离进行UPGMA聚类分析(图 3),这5个物种的10个品系的紫菜丝状体可以分成2个亚群,坛紫菜单独聚为一支;半叶紫菜、条斑紫菜、少精紫菜和皱紫菜聚为另一支,其中条斑紫菜的2个品系首先聚在一块,再与少精紫菜聚在一起,接着与皱紫菜相聚,最后与半叶紫菜汇聚。
3. 讨论
3.1 引物通用性
微卫星标记的缺点在于微卫星引物具有种属特异性,在对一个物种首次进行微卫星分析时必须要进行微卫星引物的开发,较费时耗力[10]。有关研究认为微卫星引物可以在近缘物种间进行转移应用,但能否转移以及转移的成功率在不同的物种间表现较大的差异[16-17]。SUN等[10]、刘必谦等[11]、XIE等[13]和KONG等[14]通过对一个紫菜属物种设计的微卫星引物进行了引物种间转移扩增研究,证明微卫星引物在不同物种间具有高的通用性。笔者从68对微卫星引物中筛选出20对通用微卫星引物,筛选比例为29.4%,其中14对引物均能在5个物种中进行有效扩增,证实上述选扩的微卫星位点的侧翼序列在坛紫菜、条斑紫菜、少精紫菜、半叶紫菜及皱紫菜具有比较高的保守性。
所研究的68对微卫星引物中63.24%来自紫菜EST序列,36.76%来自基因组文库,而筛选出的20对引物中有15对来自GenBank中紫菜EST数据库,占34.88%,5对来自基因组,占20%。EST序列来自于有表达意义的mRNA反转录的cDNA,相对于含有内含子的基因组文库,在物种进化中更具保守性,来自于EST序列的微卫星引物可以在更大范围的紫菜属内扩增出多态性位点。因此,EST-SSR与基因组SSR相比更容易在不同物种中进行转移扩增。
3.2 紫菜属的遗传多样性
Ne作为衡量SSR引物多态性高低的一个标准,表明了物种或品系间的遗传多样性。Ne越高,物种或品系间的遗传多样性越高,在笔者的研究中每对引物可扩增的Ne平均为2.37。张鹏等[18]对坛紫菜9个品系的亲缘关系进行SSR标记分析,平均Ne为1.72。KONG等[14]用微卫星标记对条斑紫菜进行遗传分析,平均Ne为1.81。XIE等[13]用相同标记分析了坛紫菜15品系的遗传多样性,平均Ne为2.81,此研究获得的Ne结果与前人研究相似,同时显示种间的遗传多样性高于种内。
DAYANANDAN等[19]用4个微卫星引物对就可区分白杨(Populus tremuloides)的34个品系,只有2个品系没有被区分开;BECHER等[20]用3对引物就可以很好区分44个天竺葵(Pelargonium)品系。此研究从68对微卫星引物中筛选出20对引物,多态性比例高达95.25%,这可能与引物来源有关,笔者所用的引物63.24%来自EST序列。然而,刘必谦等[7]认为从基因组DNA筛选出的微卫星,其信息量要比来自EST的丰富,而此研究结果并不支持这一观点。崔灵英等[21]用ISSR标记对4种紫菜叶状体进行分析,获得多态性条带比例达95.5%。贾建航等[22]在对15个紫菜种(系)的丝状体进行RAPD分析时得到多态性比例达97.1%;杨锐等[23]用AFLP对坛紫菜8个品系的丝状体进行分析得到96.97%的多态性位点,这些结果与笔者的研究结果相似,进一步显示了紫菜物种的遗传变异相当丰富。
3.3 5种紫菜的种间遗传关系
5个紫菜属物种10个品系丝状体的遗传相似指数平均为0.474 9,说明个体间的亲缘关系比较远。曾呈奎等[1]曾根据叶片边缘的细胞排列方式将真紫菜亚属分为全缘紫菜组、刺缘紫菜组和边缘紫菜组,并且认为该特征具有进化上的意义。根据这一分类系统皱紫菜、坛紫菜均属于刺缘紫菜组,条斑紫菜、半叶紫菜和少精紫菜属于全缘紫菜组。此研究中5个物种中少精紫菜和条斑紫菜首先聚在一起,两者的遗传距离最为接近,这一结果与崔灵英等[21]用ISSR分子标记对紫菜叶状体分析的结果一致,也符合经典分类学结果;根据笔者的研究结果,皱紫菜聚类在条斑紫菜、半叶紫菜和少精紫菜的内部,而不是与坛紫菜聚类为一支,这与杨立恩等[24]对紫菜叶状体进行rbcL基因分类分析结果相矛盾,同时此研究结果与经典形态分类学认为皱紫菜和坛紫菜都属于刺缘紫菜组的结论也不一致。造成这些差异的原因可能是:1)试验材料不一样,笔者采用的试验材料为丝状体,而其他学者为叶状体;2)各种标记手段本身也存在着差异。
前人的研究表明,坛紫菜、条斑紫菜、皱紫菜、半叶紫菜和少精紫菜这5种紫菜之间具有重要生物学特征的差异,例如少精紫菜和条斑紫菜染色体n=3,皱紫菜、半叶紫菜和坛紫菜染色体n=5[1],这一研究结果与笔者的聚类分析结果有一定的相关性。通过聚类分析结果可以较真实地反映这10个品系紫菜丝状体间的亲缘关系,进一步显示微卫星分子标记在大型海藻分类学上具有一定的应用价值。
-
-
[1] CONNELL J H. Diversity in tropical rain forests and coral reefs[J]. Science, 1978, 199(4335): 1302-1310. doi: 10.1126/science.199.4335.1302
[2] HUANG D, LICUANAN W Y, HOEKSEMA B W, et al. Extraordinary diversity of reef corals in the South China Sea[J]. Mar Biodivers, 2014, 45(2): 157-168.
[3] VERON J E N, DEVANTIER L M, TURAK E, et al. The coral triangle[M]. Dordrecht: Springer Netherlands, 2010: 47-55.
[4] BOSTRÖM-EINARSSON L, BABCOCK R C, BAYRAKTAROV E, et al. Coral restoration: a systematic review of current methods, successes, failures and future directions[J]. PLoS One, 2020, 15(1): e0226631. doi: 10.1371/journal.pone.0226631
[5] BURKE L, SELIG E, SPALDING M, et al. Reefs at risk in Southeast Asia[M]. Washington, D.C.: World Resources Institute, 2002: 2-10.
[6] 李元超, 陈石泉, 郑新庆, 等. 永兴岛及七连屿造礁石珊瑚近10年变化分析[J]. 海洋学报, 2018, 40(8): 97-109. [7] HUGHES T P, BARNES M L, BELLWOOD D R, et al. Coral reefs in the Anthropocene[J]. Nature, 2017, 546(7656): 82-90. doi: 10.1038/nature22901
[8] ANDERSON G R V, EHRLICH A H, EHRLICH P R, et al. The community structure of coral reef fishes[J]. Am Nat, 1981, 117(4): 476-495. doi: 10.1086/283729
[9] ÖHMAN M C, RAJASURIYA A. Relationships between habitat structure and fish communities on coral[J]. Environ Biol Fish, 1998, 53: 19-31. doi: 10.1023/A:1007445226928
[10] CHITTARO P M. Fish-habitat associations across multiple spatial scales[J]. Coral Reefs, 2004, 23(2): 235-244.
[11] ARIAS-GONZÁLEZ J E, DONE T J, PAGE C A, et al. Towards a reefscape ecology: relating biomass and trophic structure of fish assemblages to habitat at Davies Reef, Australia[J]. Mar Ecol Prog Ser, 2006, 320: 29-41. doi: 10.3354/meps320029
[12] CÁCERES I, ORTIZ M, CUPUL-MAGAÑA A L, et al. Trophic models and short-term simulations for the coral reefs of Cayos Cochinos and Media Luna (Honduras): a comparative network analysis, ecosystem development, resilience, and fishery[J]. Hydrobiologia, 2015, 770: 209-224.
[13] AGUILAR-MEDRANO R, CALDERON-AGUILERA L E. Redundancy and diversity of functional reef fish groups of the Mexican Eastern Pacific[J]. Mar Ecol, 2016, 37: 119-133. doi: 10.1111/maec.12253
[14] MCCLANAHAN T R, MUTHIGA N A. Changes in Kenyan coral reef community structure and function due to exploitation[J]. Hydrobiologia, 1988, 166(3): 269-276. doi: 10.1007/BF00008136
[15] KROON F J, LEFÈVRE C D, DOYLE J R, et al. DNA-based identification of predators of the corallivorous crown-of-thorns starfish (Acanthaster cf. solaris) from fish faeces and gut contents[J]. Sci Rep, 2020, 10: 8184. doi: 10.1038/s41598-020-65136-4
[16] EDWARDS C B, FRIEDLANDER A M, GREEN A G, et al. Global assessment of the status of coral reef herbivorous fishes: evidence for fishing effects[J]. Proc R Soc B, 2014, 281: 20131835. doi: 10.1098/rspb.2013.1835
[17] CRAMER K L, O'DEA A, CLARK T R, et al. Prehistorical and historical declines in Caribbean coral reef accretion rates driven by loss of parrotfish[J]. Nat Commun, 2017, 8: 14160. doi: 10.1038/ncomms14160
[18] HUGHES T P, BAIRD A H, BELLWOOD D R, et al. Climate change, human impacts, and the resilience of coral reefs[J]. Science, 2003, 301(5635): 929-933. doi: 10.1126/science.1085046
[19] HUGHES T P, RODRIGUES M J, BELLWOOD D R, et al. Phase shifts, herbivory, and the resilience of coral reefs to climate change[J]. Curr Biol, 2007, 17(4): 360-365. doi: 10.1016/j.cub.2006.12.049
[20] WILSON S K, FISHER R, PRATCHETT M S, et al. Exploitation and habitat degradation as agents of change within coral reef fish communities[J]. Glob Change Biol, 2008, 14(12): 2796-2809. doi: 10.1111/j.1365-2486.2008.01696.x
[21] NORSTRÖM A, NYSTRÖM M, LOKRANTZ J, et al. Alternative states on coral reefs: beyond coral-macroalgal phase shifts[J]. Mar Ecol Prog Ser, 2009, 376: 295-306. doi: 10.3354/meps07815
[22] 王腾, 刘永, 李纯厚, 等. 永兴岛附近海域珊瑚礁鱼类群落结构特征[J]. 水生生物学报, 2023, 47(4): 674-683. [23] 王腾, 李纯厚, 赵金发, 等. 西沙东岛珊瑚礁鱼类种类组成特征与演变[J]. 水生生物学报, 2023, 47(9): 1456-1463. doi: 10.7541/2023.2022.0448 [24] ZHAO J F, LI C H, WANG T, et al. Composition and long-term variation characteristics of coral reef fish species in Yongle Atoll, Xisha Islands, China[J]. Biology, 2023, 12(8): 1062. doi: 10.3390/biology12081062
[25] SALE P F. The ecology of fishes on coral reefs[M]. California: Academic Press, 1991: 47-51.
[26] 国家水产总局南海水产研究所, 厦门水产学院, 中国科学院海洋研究所, 等. 南海诸岛海域鱼类志[M]. 北京: 科学出版社, 1979: 1-573. [27] 王腾, 刘永, 全秋梅, 等. 七连屿珊瑚礁鱼类种类组成特征分析[J]. 中国水产科学, 2022, 29(1): 102-117. doi: 10.12264/JFSC2021-0122 [28] MOUILLOT D, LAUNE J, TOMASINI J A, et al. Assessment of coastal lagoon quality with taxonomic diversity indices of fish, zoobenthos and macrophyte communities[J]. Hydrobiologia, 2005, 550: 121-130. doi: 10.1007/s10750-005-4368-y
[29] CLARKE K, WARWICK R. A further biodiversity index applicable to species lists: variation in taxonomic distinctness[J]. Mar Ecol Prog Ser, 2001, 216: 265-278. doi: 10.3354/meps216265
[30] ZINTZEN V, ANDERSON M J, ROBERTS C D, et al. Increasing variation in taxonomic distinctness reveals clusters of specialists in the deep sea[J]. Ecography, 2011, 34(2): 306-317. doi: 10.1111/j.1600-0587.2010.06546.x
[31] ALLEN G R. Reef and shore fishes of Milne Bay Province, Papua New Guinea[C]//WERNER T B, ALLEN G R. A rapid biodiversity assessment of the coral reefs of Milne Bay Province, Papua New Guinea. RAP Working Papers 11, Washington, D.C.: Conservation International, 1988: 39-49, 67-107.
[32] 邹琦, 吴志强, 黄亮亮, 等. 广西涠洲岛珊瑚礁海域鱼类物种组成的调查分析[J]. 南方农业学报, 2020, 51(1): 1-10. doi: 10.3969/j.issn.2095-1191.2020.01.001 [33] van NGUYEN L, MAI D X. Reef fish fauna in the coastal waters of Vietnam[J]. Mar Biodivers, 2020, 50: 100. doi: 10.1007/s12526-020-01131-2
[34] KOMYAKOVA V, JONES G P, MUNDAY P L. Strong effects of coral species on the diversity and structure of reef fish communities: a multi-scale analysis[J]. PLoS One, 2018, 13(8): e0202206. doi: 10.1371/journal.pone.0202206
[35] ALLEN G R. Indo-Pacific coral-reef fishes as indicators of conservation hotspots[C]//MOOSA M K, SOEMODIHARDJO S, SOEGIARTO A, et al. Proceedings of the Ninth International Coral Reef Symposium, 23-27 October 2000, Bali, Indonesia, 2002(2): 921-926.
[36] KOESTER A, GORDÓ-VILASECA C, BUNBURY N, et al. Impacts of coral bleaching on reef fish abundance, biomass and assemblage structure at remote Aldabra Atoll, Seychelles: insights from two survey methods[J]. Front Mar Sci, 2023, 10: 1230717. doi: 10.3389/fmars.2023.1230717
[37] 雷明凤, 余克服, 廖芝衡, 等. 西沙群岛银屿珊瑚礁的生态快速退化及其对鱼类的影响[J]. 热带海洋学报, 2024, 43(3): 87-99. [38] COCHRAN K J, BOKUNIEWICZ H J, YAGER P L. Encyclopedia of ocean sciences[M]. 3rd ed. Cambridge: Academic Press, 2019: 7.
[39] CHIN A, TOBIN A, SIMPFENDORFER C, et al. Reef sharks and inshore habitats: patterns of occurrence and implications for vulnerability[J]. Mar Ecol Prog Ser, 2012, 460: 115-125. doi: 10.3354/meps09722
[40] SHERMAN C S, SIMPFENDORFER C A, PACOUREAU N, et al. Half a century of rising extinction risk of coral reef sharks and rays[J]. Nat Commun, 2023, 14: 15. doi: 10.1038/s41467-022-35091-x
[41] MCLEAN M, STUART-SMITH R D, VILLÉGER S, et al. Trait similarity in reef fish faunas across the world’s oceans[J]. Proc Nat Acad Sci, 2021, 118(12): e2012318118. doi: 10.1073/pnas.2012318118
[42] SHIN Y J, ROCHET M J, JENNINGS S, et al. Using size-based indicators to evaluate the ecosystem effects of fishing[J]. ICES J Mar Sci, 2005, 62(3): 384-396. doi: 10.1016/j.icesjms.2005.01.004
[43] CARDILLO M, MACE G M, JONES K E, et al. Multiple causes of high extinction risk in large mammal species[J]. Science, 2005, 309(5738): 1239-1241. doi: 10.1126/science.1116030
[44] HUGHES T P, KERRY J T, ÁLVAREZ-NORIEGA M, et al. Global warming and recurrent mass bleaching of corals[J]. Nature, 2017, 543(7645): 373-377. doi: 10.1038/nature21707
[45] RICHARDSON L E, GRAHAM N A J, PRATCHETT M S, et al. Structural complexity mediates functional structure of reef fish assemblages among coral habitats[J]. Environ Biol Fish, 2017, 100(3): 193-207. doi: 10.1007/s10641-016-0571-0
[46] 李嘉琪, 白爱娟, 蔡亲波. 西沙群岛和涠洲岛气候变化特征及其与近岸陆地的对比[J]. 热带地理, 2018, 38(1): 72-81. [47] MUNDAY P L, JONES G P, PRATCHETT M S, et al. Climate change and the future for coral reef fishes[J]. Fish Fish, 2008, 9(3): 261-285. doi: 10.1111/j.1467-2979.2008.00281.x
[48] HALL S, GREENSTREET S. Taxonomic distinctness and diversity measures: responses in marine fish communities[J]. Mar Ecol Prog Ser, 1998, 166: 227-229. doi: 10.3354/meps166227
[49] ROGERS S I, CLARKE K R, REYNOLDS J D. The taxonomic distinctness of coastal bottom-dwelling fish communities of the Northeast Atlantic[J]. J Anim Ecol, 1999, 68(4): 769-782. doi: 10.1046/j.1365-2656.1999.00327.x
[50] CAMPBELL N, NEAT F, BURNS F, et al. Species richness, taxonomic diversity, and taxonomic distinctness of the deep-water demersal fish community on the Northeast Atlantic continental slope (ICES Subdivision VIa)[J]. ICES J Mar Sci, 2010, 68(2): 365-376.
[51] CHAUDHARY C, RICHARDSON A J, SCHOEMAN D S, et al. Global warming is causing a more pronounced dip in marine species richness around the equator[J]. Proc Natl Acad Sci, 2021, 118(15): e2015094118. doi: 10.1073/pnas.2015094118
[52] PANKHURST N W, MUNDAY P L. Effects of climate change on fish reproduction and early life history stages[J]. Mar Freshw Res, 2011, 62(9): 1015. doi: 10.1071/MF10269
[53] 李媛洁, 陈作志, 张俊, 等. 西沙群岛七连屿礁栖鱼类物种和分类多样性[J]. 中国水产科学, 2020, 27(7): 815-823. [54] JIANG X M, PAN B Z, SUN Z W, et al. Application of taxonomic distinctness indices of fish assemblages for assessing effects of river-lake disconnection and eutrophication in floodplain lakes[J]. Ecol Indic, 2020, 110: 105955. doi: 10.1016/j.ecolind.2019.105955
[55] DÍAZ S, FARGIONE J, CHAPIN F S, et al. Biodiversity loss threatens human well-being[J]. PLoS Biol, 2006, 4(8): e277. doi: 10.1371/journal.pbio.0040277
[56] DULVY N K, SADOVY Y, REYNOLDS J D. Extinction vulnerability in marine populations[J]. Fish Fish, 2003, 4(1): 25-64. doi: 10.1046/j.1467-2979.2003.00105.x
[57] HIDDINK J G, MACKENZIE B R, RIJNSDORP A, et al. Importance of fish biodiversity for the management of fisheries and ecosystems[J]. Fish Res, 2008, 90(1/2/3): 6-8.
-
其他相关附件
-
PDF格式
20240008附录A 点击下载(263KB)
-



 下载:
下载:



 粤公网安备 44010502001741号
粤公网安备 44010502001741号
