Comparison between single factor design and orthogonal design to optimize reaction system for TRAP markers in tilapia
-
摘要:
靶位区域扩增多态性(target region amplification polymorphism,TRAP)是一种新型功能性分子标记,比较了L16(45)正交试验和单因素方法优化罗非鱼(Oreochromis spp.)TRAP反应体系。2种分析方法结果中dNTPs浓度、随机引物浓度、DNA浓度相同而镁离子(Mg2+)浓度和TaqDNA聚合酶浓度不同,不同因素间存在一定的交互作用。正交设计方法比单因素法更简单、科学、合理。该试验获得罗非鱼15 μL TRAP最优反应体系为DNA模板浓度为60 ng、Mg2+浓度为1.5 mmol·L-1、dNTPs浓度为0.3 mmol·L-1、TaqDNA聚合酶0.5 U、随机引物浓度为7 pmol·L-1和固定引物浓度为10 pmol·L-1。该反应体系在4个罗非鱼群体中验证,表现出良好的稳定性和重复性,该反应体系的建立为TRAP标记应用于罗非鱼遗传多样性评价、种质鉴定、分子标记辅助育种等研究提供了新的手段。
Abstract:The target region amplification polymorphism (TRAP) marker is a novel functional molecular marker. In present study, we compared the optimization of the reaction system for TRAP markers in tilapia (Oreochromis spp.) between L16(45) orthogonal design and single-factor design. The results of the two approaches showed that the concentrations of dNTPs, random primer and DNA were the same, while those of Mg2 + and TaqDNA polymerase dosage varied, and interaction among different factors were observed. The orthogonal design is simpler, more scientific and reasonable than single factor design. The optimal TRAP reaction system for tilapia includes 60 ng DNA template, 1.5 mmol·L-1 Mg2+, 0.3 mmol·L-1 dNTPs, 0.5 U TaqDNA polymerase, 7 pmol·L-1 random primers and 10 pmol·L-1 fixed primer. The stability and repeatability of the reaction system had been verified in 4 tilapia populations. The TRAP marker provides new insights into evaluation of genetic diversity, germplasm identification, molecular marker-assisted breeding and other related researches of tilapia.
-
Keywords:
- tilapia /
- TRAP /
- orthogonal design /
- single-factor test /
- optimization
-
罗非鱼(Oreochromis spp.)属鲈形目,丽鱼科,是原产于非洲的热带性鱼类,得到联合国粮农组织的大力推荐,成为世界最重要的养殖鱼类之一[1-2]。2010年罗非鱼全球产量达到320×104 t,中国占其35%,居于世界首位[3-4]。当前实际生产中存在较多的罗非鱼种间杂交,从而使得种质发生退化和混杂,因此迫切需要寻找群体间的遗传标记[5]用于种质鉴定。近年来广泛应用的DNA分子标记在罗非鱼等水产动物的遗传多样性、近亲繁殖、品系鉴定等方面都有很大进展[6]。DNA分子标记由于不受环境、季节、发育阶段、组织的影响与限制,能够直接反应DNA的多态性,易于建立与目标性状的关联,其在鱼类种质鉴定、遗传多样性分析、遗传图谱构建、基因定位、亲缘关系分析、分子辅助育种中得到广泛的研究与应用[7-8]。其中基于保守序列、保守基序或保守调控核心序列或真核生物中基因序列的特点,以及表达序列标签(EST)的目标分子标记技术受到越来越多研究者的重视[9-10]。基于基因序列和EST的靶位区域扩增多态性(target region amplification polymorphism,TRAP)是HU和VICK[11]在向日葵(Helianthus annuus)上提出的新型目标分子标记,其利用EST或cDNA序列信息和生物信息工具,对目标区域序列进行扩增产生多态性分子标记。TRAP具有操作简单、重复性好、效率高和易测序等优点[12],随后在水稻(Oryza sativa)、莴苣(Lactuca sativa)、小麦(Triticum aestivum)、油菜(Brassica napus)等植物的遗传多样性分析、重要性状标记筛选、遗传图谱构建等得到应用[13-17]。同时TRAP在草鱼(Ctenopharyngodon idella)、鲤(Cyprinus carpio)、鲶(Silurus asotus)、鳜(Siniperca chuatsi)等水产动物研究中也得到运用[18-20]。但TRAP在罗非鱼上的应用以及应用过程中的PCR反应体系的建立未见报道。最早由何正文等[21]提出的正交设计法优化PCR反应体系方法,由于其试验规模小且能够考虑各因素间的交互作用,随后用于大麻哈鱼(Oncorhynchus keta)SRAR-PCR、葡萄(Vitis vinifera)SCoT-PCR等PCR反应体系优化中[22-23]。而水产动物TRAP反应体系优化都是以保持体系中其他参数不变针对一个影响因子浓度进行优化的单因素法,而未见正交设计法的使用[19, 24]。
笔者比较了正交试验设计和单因素分析法,建立了稳定的罗非鱼TRAP反应体系,以期为罗非鱼的遗传多样性分析、种质鉴定、分子辅助育种等提供新的研究手段。
1. 材料与方法
1.1 材料
试验所用4个罗非鱼群体均来自中国水产科学研究院淡水渔业研究中心宜兴养殖基地,分别是马来西亚红罗非鱼(O.niloticus)、奥尼罗非鱼(O.niloticus×O.aureus)、吉富罗非鱼(O.niloticus Gift strain)和新吉富罗非鱼(O.niloticus New Gift strain)。
1.2 DNA提取与检测
每个群体随机取20个个体,剪尾鳍于95%的酒精中固定并在-20 ℃冰箱保存。尾鳍在加入液氮的研钵中充分研磨后参照Takara的universal genomic DNA extraction kit Ver 3.0试剂盒中的动物组织提取基因组方法提取DNA。所得DNA用1%琼脂糖凝胶电泳检测质量并用分光光度计检测其浓度与纯度,稀释适当浓度后于-20 ℃保存。
1.3 TRAP-PCR扩增
固定引物序列参考HU和VICK[11]设计的4个固定引物,随机引物参考HU[25]的6个随机引物(表 1),引物由Invitrogen Biotechnology公司合成。DNA扩增程序参考HU和VICK[11]等,略改动为94 ℃ 2 min,94 ℃ 45 s,38 ℃ 45 s,72 ℃ 1 min,5个循环;94 ℃ 45 s,53 ℃ 45 s,72 ℃ 1 min,35个循环;72 ℃延伸8 min,最后于4 ℃保存。
表 1 试验采用的4个固定引物和6个随机引物序列Table 1. Sequences of 4 fixed primers and 6 arbitrary primers used in present study引物 primer 序列 sequence (5′→3′) 固定引物fixed primer E1 AGATTTCACCAAGGCTGT E2 TTATGAATTGCTGGCTTG E3 TGAATCCGTCTTCATTCA E4 ATCGATGCCTTTGATTTT 随机引物arbitrary primer Ga3-800 TCATCTCAAACCATCTACAC Odd26-700 CTATCTCTCGGGACCAAAC Sa12-700 TTCTAGGTAATCCAACAACA Trap03-700 CGTAGCGCGTCAATTATG Trap13-800 GCGCGATGATAAATTATC Em1 GGAACCAAACACATGAAGA 1.4 TRAP产物检测
PCR扩增结束后在300 V电压下用8%聚丙烯酰胺凝胶电泳3 h,用DNAGREEN绿如蓝低毒核酸染料(北京天恩泽基因科技有限公司出品)染色15 min,由美国UVP GelDoc-ItTM imaging system凝胶成像系统拍照保存。
1.5 试验设计
1.5.1 正交设计
以红罗非鱼DNA为模板筛选引物,选择一对较好的引物进行扩增,将影响罗非鱼TRAP反应体系的镁离子(Mg2+)浓度、dNTPs浓度、TaqDNA聚合酶浓度、模板DNA浓度和引物浓度5个因子设置4个水平,用L16(45)正交试验对罗非鱼TRAP反应体系进行分析、优化(表 2),3次重复。然后依据条带灵敏性、特异性及清晰度作1~16分打分[21],运用正交设计软件助手进行结果分析。
表 2 红罗非鱼TRAP反应体系L16(45)正交设计Table 2. L16(45) orthogonal design of red tilapia TRAP reaction system编号
No.DNA模板/ng
DNA template镁离子/mmol·L-1
Mg2+脱氧核苷酸/mmol·L-1
dNTPs随机引物/pmol·L-1
arbitrary primerTaqNDA聚合酶/U
Taq NDA polymerase1 30 1.0 0.20 0.5 0.3 2 30 1.5 0.25 2.0 0.5 3 30 2.5 0.30 7.0 0.8 4 30 3.0 0.35 10.0 1.1 5 60 1.0 0.25 7.0 1.1 6 60 1.5 0.20 10.0 0.8 7 60 2.5 0.35 0.5 0.5 8 60 3.0 0.30 2.0 0.3 9 90 1.0 0.30 10.0 0.5 10 90 1.5 0.35 7.0 0.3 11 90 2.5 0.20 2.0 1.1 12 90 3.0 0.25 0.5 0.8 13 120 1.0 0.35 2.0 0.8 14 120 1.5 0.30 0.5 1.1 15 120 2.5 0.25 10.0 0.3 16 120 3.0 0.20 7.0 0.5 1.5.2 单因素对比试验
参考HU和VICK[11]15 μL TRAP反应体系中的各参数,用DNA模板50 ng,Mg2+ 2.5 mmol·L-1、dNTPs 0.3 mmol·L-1、TaqDNA聚合酶1.5 U、固定引物10 pmol·L-1(随机引物浓度/固定引物浓度为1/1)和1×PCR buffer(Mg2+ Free)。针对某一因素设计不同浓度处理,而其他因素浓度不变。固定引物与buffer浓度不变,提高固定引物与随机引物的比例[26]。最后对正交设计和单因素法优化效果进行比较。
2. 结果与分析
2.1 正交设计
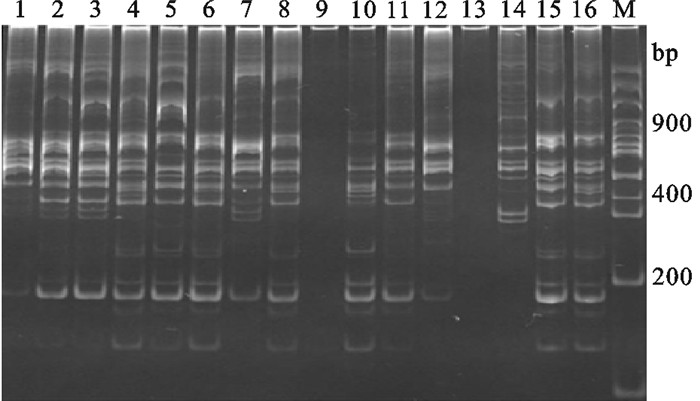
罗非鱼TRAP正交设计试验结果表明,Mg2+、dNTPs、TaqDNA聚合酶、模板DNA和引物等不同浓度混合进行PCR扩增,可产生显著差异的条带多态性以及条带清晰度(图 1)。其中9和13无扩增结果,泳道1杂带很多且难区分;3和7条带较亮且清晰,最佳处理组合为3。用正交设计软件助手对16个反应组合进行直观分析和方差分析,结果见图 2和表 3。
表 3 罗非鱼TRAP反应体系正交设计方差分析Table 3. Variance analysis of TRAP reaction system for tilapia optimized with orthogonal design变异来源
source极差
range方差
SSF 均方
MS标准差
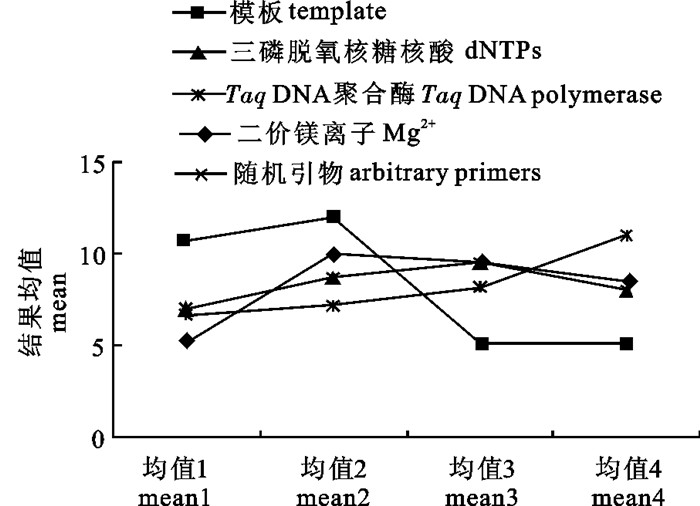
X±SD模板 template 6.750 153.188 2.428 51.063 12.37 二价镁离子 Mg2+ 4.750 54.688 0.867 18.229 7.39 三磷脱氧核糖核酸 dNTPs 2.500 13.688 0.217 4.563 3.70 随机引物 arbitrary primer 4.750 50.688 0.803 16.896 7.12 TaqDNA聚合酶 TaqDNA 4.250 43.188 0.685 14.396 6.57 共计 total 315.440 均值大小代表各因素各水平间的差异,而极差大小代表该因素对试验的影响程度[22]。图 2中DNA不同浓度均值变化最大,影响最显著,其余依次为Mg2+、TaqDNA聚合酶、dNTPs和随机引物。表 3可看出模板影响最为显著。根据DNA浓度各水平均值, 显示条带并非浓度越高扩展扩增效果更好,60 ng时显著高于其他浓度水平,扩增条带较清晰丰富。dNTPs、随机引物和Mg2+浓度变化均值均出现了波峰,由此可见PCR反应各因子浓度均应控制在某一范围,扩增效果与各因子浓度不相关。综合各因素得到罗非鱼15 μL TRAP正交试验最优反应体系为DNA模板浓度为60 ng、Mg2+浓度为1.5 mmol·L-1、dNTPs浓度为0.3 mmol·L-1、TaqDNA聚合酶0.5 U和随机引物浓度为7 pmol·L-1。
2.2 单因素试验
2.2.1 dNTPs浓度、随机引物浓度、DNA浓度对罗非鱼TRAP扩增效果的影响
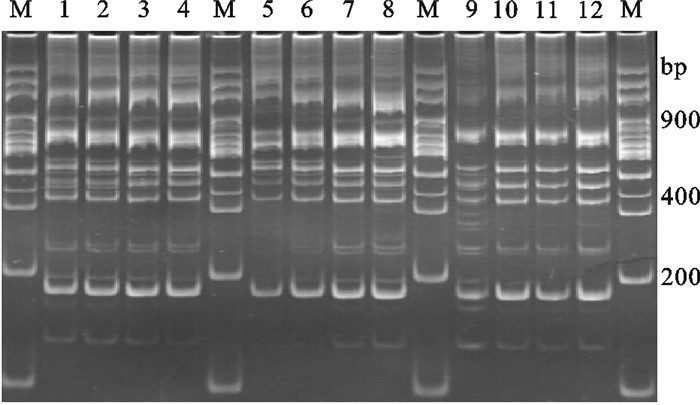
dNTPs浓度、随机引物浓度和DNA浓度对罗非鱼TRAP扩增效果的影响见图 3。结果显示,dNTPs浓度增加,特异性条带亮度增加;浓度较低时杂带较多,条带相对不够清晰。罗非鱼15 μL TRAP反应体系中,dNTPs浓度在0.3 mmol·L-1时条带亮度、清晰度以及特异性高于其他水平。当随机引物浓度增加时条带清晰且丰富;浓度较低时条带较少且不太清晰,但随机引物浓度为7 pmol·L-1时条带清晰度和丰富性最好。当DNA浓度较低时杂带较多且很模糊,罗非鱼15 μL TRAP反应体系中当DNA浓度达到60 ng以上,扩增效果没有明显差异。
![]() 图 3 dNTPs浓度、随机引物浓度和DNA浓度对罗非鱼TRAP的影响M. 100 bp DNA ladder;1~12. dNTPs浓度、随机引物浓度和DNA浓度各自1~4处理水平Figure 3. Effects of concentrations of dNTPs, arbitrary primers and template on TRAP reaction system for tilapiaM. 100 bp DNA ladder; 1~12. 1~4 levels of concentrations of dNTPs, random primers and template
图 3 dNTPs浓度、随机引物浓度和DNA浓度对罗非鱼TRAP的影响M. 100 bp DNA ladder;1~12. dNTPs浓度、随机引物浓度和DNA浓度各自1~4处理水平Figure 3. Effects of concentrations of dNTPs, arbitrary primers and template on TRAP reaction system for tilapiaM. 100 bp DNA ladder; 1~12. 1~4 levels of concentrations of dNTPs, random primers and template2.2.2 Mg2+浓度和TaqDNA聚合酶浓度对罗非鱼TRAP扩增效果的影响
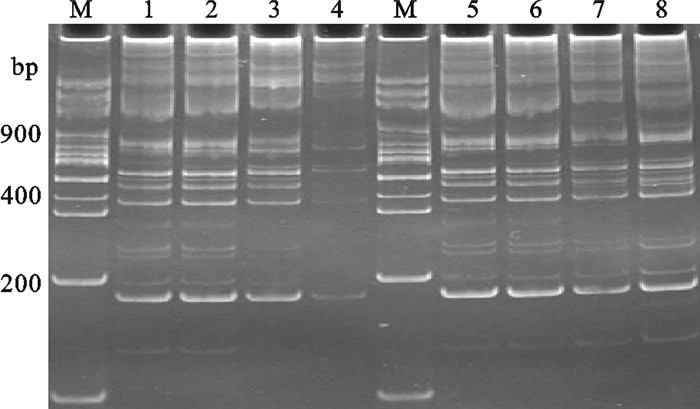
Mg2+浓度较低时产生的条带较少且不清晰(图 4)。罗非鱼15 μL TRAP反应体系中,Mg2+浓度为2.5 mmol·L-1时其条带清晰度与亮度明显高于其他3个浓度水平。TaqDNA聚合酶在所选试验浓度范围内浓度较高时条带较清晰,但5和6条带质量差距不大,综合考虑成本和条带质量因素,罗非鱼15 μL TRAP反应体系TaqDNA聚合酶浓度在0.8 U时较好。运用单因素分析法得到罗非鱼15 μL TRAP最优反应体系为DNA模板浓度为60 ng、Mg2+ 浓度为2.5 mmol·L-1、dNTPs浓度为0.3 mmol·L-1、TaqDNA聚合酶浓度为0.8 U和随机引物浓度为7 pmol·L-1。
2.3 TRAP反应体系2种优化方法比较及验证
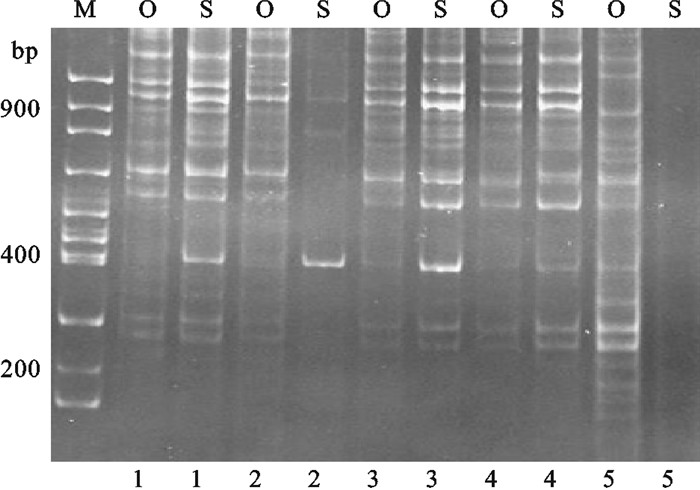
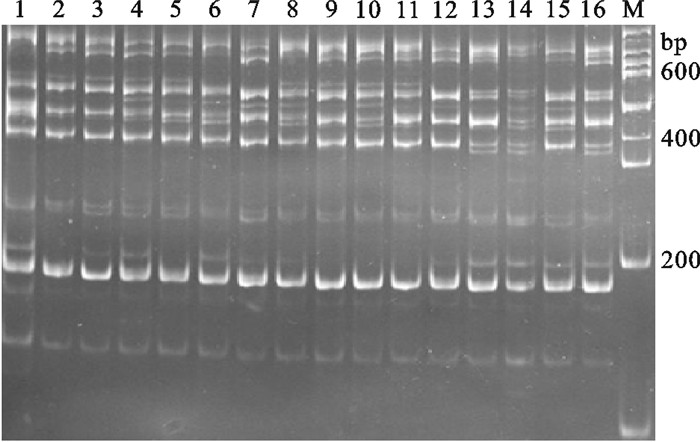
用正交设计和单因素方法分别优化罗非鱼TRAP反应体系的结果,对随机挑取的5个红罗非鱼个体PCR扩增对比,单因素优化罗非鱼TRAP-PCR产生的条带正交设计法优化结果几乎都能够产生,但正交设计法优化结果能扩增出较多单因素法优化结果无法扩增出的条带(图 5)。以4个罗非鱼群体材料对优化后体系进行验证,扩增条带清晰且特异性强,特异性条带较多,说明此反应体系适于罗非鱼TRAP-PCR分析(图 6)。
![]() 图 5 比较正交设计和单因素优化罗非鱼TRAP扩增图M. 100 bp DNA ladder;O. 正交设计;S. 单因素;1~5. 不同罗非鱼个体Figure 5. Comparison of amplification results of TRAP reaction system for tilapia between orthogonal design and single factor designM. 100 bp of DNA ladder; O. orthogonal design; S. single factor; 1~5. different tilapia individuals
图 5 比较正交设计和单因素优化罗非鱼TRAP扩增图M. 100 bp DNA ladder;O. 正交设计;S. 单因素;1~5. 不同罗非鱼个体Figure 5. Comparison of amplification results of TRAP reaction system for tilapia between orthogonal design and single factor designM. 100 bp of DNA ladder; O. orthogonal design; S. single factor; 1~5. different tilapia individuals![]() 图 6 4个不同罗非鱼群体的TRAP扩增图M. 100 bp DNA ladder;1~4. 马来西亚红罗非鱼;5~8. 新吉富罗非鱼;9~12. 吉富罗非鱼;13~16. 奥尼罗非鱼Figure 6. Amplification result of TRAP reaction system for 4 different tilapia populationsM. 100 bp DNA ladder; 1~4. red tilapia; 5~8. New GIFT strain Nile tilapia; 9~12. GIFT strain Nile tilapia; 13~16. O.niloticus×O.aureus
图 6 4个不同罗非鱼群体的TRAP扩增图M. 100 bp DNA ladder;1~4. 马来西亚红罗非鱼;5~8. 新吉富罗非鱼;9~12. 吉富罗非鱼;13~16. 奥尼罗非鱼Figure 6. Amplification result of TRAP reaction system for 4 different tilapia populationsM. 100 bp DNA ladder; 1~4. red tilapia; 5~8. New GIFT strain Nile tilapia; 9~12. GIFT strain Nile tilapia; 13~16. O.niloticus×O.aureus3. 讨论
在罗非鱼等水产动物中应用比较广的遗传标记主要是限制性片段长度多态性(restriction fragment length polymorphism,RFLP)、随机扩增多态性DNA(random amplified polymorphic RAPD)、扩增片段长度多态性(amplified fragment length polymorphism,AFLP)、单核苷酸的多态性(single nucleotide polymorphisms,SNP)、微卫星标记(simple sequence repeat,SSR)和表达序列标签(expressed sequence tag,EST)等[7, 27-28]。但随着大规模DNA测序技术的迅速发展,很多重要经济鱼类都获得了大量的DNA序列信息。分子标记技术也伴随着DNA测序技术发展逐步从传统的匿名性分子标记转向功能性及目标性分子标记[5]。TRAP标记与RAPD一样具有易于建立、操作、重复性强等特点,同时由于其特定引物中的固定引物可来自功能基因序列,所以具有能够偏向产生功能基因或者目标性区域条带的优势。TRAP能够把EST或cDNA序列与动物生长性状建立联系,能够快速筛选出生长相关的分子标记。TRAP标记既具有AFLP技术强大功能的优势,同时具有简单、多态性丰富、效率高、稳定性好的特点。其已经成功用于鲤、草鱼、鲶和鳜等水产动物的遗传多样分析以及分子标记辅助育种等[18-20, 29]。目前对于鲤、草鱼、棉花、水稻等[16, 19, 24, 26]TRAP反应体系的建立与优化,都是运用单因素分析法。单因素分析法是对每个反应影响因子进行逐项优化,不同因素、不同水平产生的反应结果对比鲜明,易于挑选单一因素最佳水平。但其不能够充分考虑单因素间的交互作用,不能保证各因素最佳浓度时组合为最佳反应组合。而运用统计学方法的正交设计法能够考察各因素间的交互作用,利用正交表安排试验方案,能够在最少的试验次数中获得最佳结果,一般各因素最好水平组合就是最优方案,但此方法具有一定局限性[22],如根据条带的清晰度和特异性对条带进行打分,具有一定的主观性。因此,该试验同时采用单因素分析法和正交设计法比较进行优化。
正交设计分析法和单因素分析法优化罗非鱼TRAP反应体系结果表明,2种分析方法结果为dNTPs浓度、随机引物浓度、DNA浓度相同,Mg2+浓度和TaqDNA聚合酶浓度不相同,两者在Mg2+浓度和TaqDNA聚合酶浓度对PCR扩增清晰度和多态性具有差异,在一定浓度范围内Mg2+浓度和TaqDNA聚合酶浓度与其他因子互作对PCR扩增具有显著性交互作用,可能是正交设计考虑了单因素间的交互作用。酶浓度超过一定范围对于PCR扩增效果没有显著作用,反而会引起非特异性扩增,因此,应比较分析2种方法中各因素使用浓度。该试验对正交设计与单因素法优化结果比较,正交设计法优化结果产生更多的多态性条带,因其能够综合考虑罗非鱼TRAP-PCR反应各因素之间的相互作用。该试验优化是建立在对其他鱼类TRAP反应各因子浓度有一定了解的基础上,如果对各因子浓度范围不清楚,可用正交设计试验探寻每个因素大概适宜浓度,然后再利用单因素分析法的快捷、对比鲜明等特点对PCR反应因子进行逐步优化[22]。为了获得稳定且重复性好的TRAP反应体系或是产生更加偏向靶位区域的条带,在草鱼、向日葵[24, 25]的TRAP反应体系都提高了起始退火温度到38 ℃和40 ℃,笔者试验亦采用38 ℃。罗非鱼TRAP反应体系中固定引物浓度与随机引物浓度之比与草鱼、鲶等[19, 23]均不相同而与鲤相似[19]。罗非鱼与草鱼、鲶染色体数量相似,但与鲤差距较大[30],可见不同鱼种的反应体系的差异与染色体的数量没有直接关系,可能与基因组核苷酸序列或空间结构相关,所以不同种鱼间的TRAP反应体系各有不同,不能简单套用。此外,该试验采用DNAGREEN对扩增产物染色,其具有EB相当的稳定性和灵敏度,但毒性较低。
综合单因素与正交设计试验能够获得更科学、完善的试验结果[23]。比较2种分析方法,正交设计方法比单因素法更简单、科学、合理。笔者试验建立反应稳定、重复性好的罗非鱼15μL TRAP最优反应体系是DNA模板浓度为60 ng、Mg2+浓度为1.5 mmol·L-1、dNTPs浓度为0.3 mmol·L-1、TaqDNA聚合酶0.5 U、随机引物浓度为7 pmol·L-1和固定引物浓度为10 pmol·L-1。该TRAP反应体系均能在马来西亚红罗非鱼、新吉富罗非鱼、吉富罗非鱼、奥尼罗非鱼群体间和群体中扩增出多态性较好的条带,大小主要在100~900 bp。该反应体系的建立为罗非鱼遗传多样性评价、种质鉴定、分子标记辅助育种等研究提供了新的手段。
-
图 3 dNTPs浓度、随机引物浓度和DNA浓度对罗非鱼TRAP的影响
M. 100 bp DNA ladder;1~12. dNTPs浓度、随机引物浓度和DNA浓度各自1~4处理水平
Figure 3. Effects of concentrations of dNTPs, arbitrary primers and template on TRAP reaction system for tilapia
M. 100 bp DNA ladder; 1~12. 1~4 levels of concentrations of dNTPs, random primers and template
图 5 比较正交设计和单因素优化罗非鱼TRAP扩增图
M. 100 bp DNA ladder;O. 正交设计;S. 单因素;1~5. 不同罗非鱼个体
Figure 5. Comparison of amplification results of TRAP reaction system for tilapia between orthogonal design and single factor design
M. 100 bp of DNA ladder; O. orthogonal design; S. single factor; 1~5. different tilapia individuals
图 6 4个不同罗非鱼群体的TRAP扩增图
M. 100 bp DNA ladder;1~4. 马来西亚红罗非鱼;5~8. 新吉富罗非鱼;9~12. 吉富罗非鱼;13~16. 奥尼罗非鱼
Figure 6. Amplification result of TRAP reaction system for 4 different tilapia populations
M. 100 bp DNA ladder; 1~4. red tilapia; 5~8. New GIFT strain Nile tilapia; 9~12. GIFT strain Nile tilapia; 13~16. O.niloticus×O.aureus
表 1 试验采用的4个固定引物和6个随机引物序列
Table 1 Sequences of 4 fixed primers and 6 arbitrary primers used in present study
引物 primer 序列 sequence (5′→3′) 固定引物fixed primer E1 AGATTTCACCAAGGCTGT E2 TTATGAATTGCTGGCTTG E3 TGAATCCGTCTTCATTCA E4 ATCGATGCCTTTGATTTT 随机引物arbitrary primer Ga3-800 TCATCTCAAACCATCTACAC Odd26-700 CTATCTCTCGGGACCAAAC Sa12-700 TTCTAGGTAATCCAACAACA Trap03-700 CGTAGCGCGTCAATTATG Trap13-800 GCGCGATGATAAATTATC Em1 GGAACCAAACACATGAAGA 表 2 红罗非鱼TRAP反应体系L16(45)正交设计
Table 2 L16(45) orthogonal design of red tilapia TRAP reaction system
编号
No.DNA模板/ng
DNA template镁离子/mmol·L-1
Mg2+脱氧核苷酸/mmol·L-1
dNTPs随机引物/pmol·L-1
arbitrary primerTaqNDA聚合酶/U
Taq NDA polymerase1 30 1.0 0.20 0.5 0.3 2 30 1.5 0.25 2.0 0.5 3 30 2.5 0.30 7.0 0.8 4 30 3.0 0.35 10.0 1.1 5 60 1.0 0.25 7.0 1.1 6 60 1.5 0.20 10.0 0.8 7 60 2.5 0.35 0.5 0.5 8 60 3.0 0.30 2.0 0.3 9 90 1.0 0.30 10.0 0.5 10 90 1.5 0.35 7.0 0.3 11 90 2.5 0.20 2.0 1.1 12 90 3.0 0.25 0.5 0.8 13 120 1.0 0.35 2.0 0.8 14 120 1.5 0.30 0.5 1.1 15 120 2.5 0.25 10.0 0.3 16 120 3.0 0.20 7.0 0.5 表 3 罗非鱼TRAP反应体系正交设计方差分析
Table 3 Variance analysis of TRAP reaction system for tilapia optimized with orthogonal design
变异来源
source极差
range方差
SSF 均方
MS标准差
X±SD模板 template 6.750 153.188 2.428 51.063 12.37 二价镁离子 Mg2+ 4.750 54.688 0.867 18.229 7.39 三磷脱氧核糖核酸 dNTPs 2.500 13.688 0.217 4.563 3.70 随机引物 arbitrary primer 4.750 50.688 0.803 16.896 7.12 TaqDNA聚合酶 TaqDNA 4.250 43.188 0.685 14.396 6.57 共计 total 315.440 -
[1] MURTHY L N, PANDA S K, SHAMAUNDAR B A. Physico-chemical and functional properties of proteins of tilapia (Oreochromis mossambicus)[J]. J Food Process Engin, 2011, 34(1): 83-107. doi: 10.1111/j.1745-4530.2008.00338.x
[2] 卢迈新, 黄樟翰. 罗非鱼遗传育种研究[J]. 上海水产大学学报, 2005, 14(2): 186-191. doi: 10.3969/j.issn.1004-7271.2005.02.016 [3] THODESEN J, RYE M, WANG Y X, et al. Genetic improvement of tilapias in China: genetic parameters and selection responses in growth of Nile tilapia (Oreochromis niloticus) after six generations of multi-trait selection for growth and fillet yield[J]. Aquaculture, 2011, 322/323: 51-64. doi: 10.1016/j.aquaculture.2011.10.010
[4] 储霞玲, 曹俊明, 白雪娜, 等. 2011年广东罗非鱼产业发展现状分析[J]. 广东农业科学, 2012(8): 12-14. doi: 10.3969/j.issn.1004-874X.2009.07.083 [5] 杜诚, 卢迈新, 叶星, 等. 5个罗非鱼群体的遗传分析与分子标记[J]. 大连水产学院学报, 2005, 20(1): 25-28. https://xueshu.baidu.com/usercenter/paper/show?paperid=1b5k02g03c4e00d01j2p06v0e6236380&site=xueshu_se&hitarticle=1 [6] 王忠华, 董西征, 钱国英. DNA分子标记技术在水生动物遗传多样性研究中的应用[J]. 科技通报, 2008, 24(5): 623-630. doi: 10.3969/j.issn.1001-7119.2008.05.007 [7] GOLDBERG D, JACKSON K, YEHUDA Y, et al. Application of RAPD in the study of genetic variations between African and American Cichlidae[J]. Indian J Fish, 2011, 46(3): 307-312. https://xueshu.baidu.com/usercenter/paper/show?paperid=83bc4f7119b155e5964a745269b67cae&site=xueshu_se
[8] CHIU T H, SU Y C, PAI J Y, et al. Molecular markers for detection and diagnosis of the giant grouper (Epinephelus lanceolatus)[J]. Food Control, 2011, 24(1/2): 29-37. https://www.sciencedirect.com/science/article/abs/pii/S0956713511003549
[9] 熊发前, 蒋菁, 钟瑞春, 等. 分子标记技术的两种新分类思路及目标分子标记技术的提出[J]. 中国农学通报, 2010, 26(10): 60-64. [10] 姚红伟, 张立冬, 孙金阳, 等. DNA分子标记技术概述[J]. 河北渔业, 2010(7): 42-46. doi: 10.3969/j.issn.1004-6755.2010.07.020 [11] HU J, VICK B. Target region amplification polymorphism: a novel marker technique for plant genotyping[J]. Plant Mol Biol Rep, 2003, 21(3): 289-294. doi: 10.1007/BF02772804
[12] 边银丙, 宋小亚. 几种新型DNA分子标记及其在食用菌研究中的应用[J]. 食用菌学报, 2006, 13(1): 78-87. doi: 10.3969/j.issn.1005-9873.2006.01.010 [13] LIU Z H, ANDERSON J A, HU J, et al. A wheat intervarietal genetic linkage map based on microsatellite and target region amplified polymorphism markers and its utility for detecting quantitative trait loci[J]. Theor Appl Genet, 2005, 111(4): 782-794. doi: 10.1007/s00122-005-2064-y
[14] HU J, OCHOA O E, TRUCO M J, et al. Application of the TRAP technique to lettuce (Lactuca sativa L. ) genotyping[J]. Euphytica, 2005, 144(3): 225-235. doi: 10.1007/s10681-005-6431-1
[15] MIKLAS P N, HU J, GRUNWALD N J, et al. Potential application of TRAP (targeted region amplified polymorphism) markers for mapping and tagging disease resistance traits in common bean[J]. Crop Sci, 2006, 46(2): 910-916. doi: 10.2135/cropsci2005.08-0242
[16] 朱志凯, 方良俊, 招倩婷, 等. 水稻TRAP-PCR反应体系优化与P-糖蛋白基因片段的分析[J]. 分子植物育种, 2008, 6(1): 65-70. doi: 10.3969/j.issn.1672-416X.2008.01.010 [17] AL-DOSS A A, ELSHAFEI A A, MOUSTAFA K A, et al. Comparative analysis of diversity based on morpho-agronomic traits and molecular markers in durum wheat under heat stress[J]. Afr J Biotechnol, 2011, 10(19): 3671-3681. doi: 10.1007/s10681-009-9965-9
[18] 梅秋兰. 鳜原种群体和养殖群体遗传多样性的TRAP及SSR标记分析[D]. 长沙: 湖南农业大学, 2010. https://cstj.cqvip.com/Qikan/Article/Detail?id=33823794&from=Qikan_Article_Detail [19] 曲疆奇, 苏胜彦, 董在杰, 等. 鲤TRAP分子标记反应体系的建立与优化[J]. 动物学杂志, 2010, 45(3): 72-78. https://cstj.cqvip.com/Qikan/Article/Detail?id=34103419&from=Qikan_Search_Index [20] 葛学亮, 孙中武, 尹洪滨. 人工养殖鲶的TRAP遗传分析[J]. 大连水产学院学报, 2010, 25(1): 76-79. https://cstj.cqvip.com/Qikan/Article/Detail?id=33030070&from=Qikan_Search_Index [21] 何正文, 刘运生, 陈立华, 等. 正交设计直观分析法优化PCR条件[J]. 湖南医科大学学报, 1998(4): 76-77. https://cstj.cqvip.com/Qikan/Article/Detail?id=3168505&from=Qikan_Search_Index [22] 张君玉, 郭大龙, 龚莹, 等. 葡萄目标起始密码子多态性反应体系的优化[J]. 果树学报, 2011(2): 209-214. https://cstj.cqvip.com/Qikan/Article/Detail?id=37026487&from=Qikan_Search_Index [23] 张慧, 孙中武, 刘伟, 等. 大麻哈鱼SRAP反应体系正交设计及优化[J]. 水产学杂志, 2010, 23(2): 6-10. https://cstj.cqvip.com/Qikan/Article/Detail?id=34491823&from=Qikan_Search_Index [24] 张志伟, 韩曜平, 曹哲明. 草鱼TRAP-PCR反应体系的建立[J]. 生物技术, 2006, 16(2): 31-34. https://cstj.cqvip.com/Qikan/Article/Detail?id=21699731&from=Qikan_Search_Index [25] HU J. Defining the sunflower (Helianthus annuus L. ) linkage group ends with the Arabidopsis-type telomere sequence repeat-derived markers[J]. Chromosome Res, 2006, 14(5): 535-548.
[26] 苗培明, 罗淑萍, 范玲. 棉花TRAP-PCR扩增体系的建立[J]. 新疆农业科学, 2009, 45(6): 1007-1011. https://cstj.cqvip.com/Qikan/Article/Detail?id=28921460&from=Qikan_Search_Index [27] CHAUHAN T, RAJIV K. Molecular markers and their applications in fisheries and aquaculture[J]. Adv Biosci Biotechnol, 2010, 1(4): 281-291. doi: 10.4236/abb.2010.14037
[28] 李莉好, 喻达辉, 黄桂菊, 等. 吉富罗非鱼不同选育群体的遗传多样性[J]. 南方水产, 2007, 3(5): 40-48. https://cstj.cqvip.com/Qikan/Article/Detail?id=27484867&from=Qikan_Search_Index [29] 董在杰, 曲疆奇, 袁新华, 等. 鲤改良品系与体质量、体长性状相关的TRAP标记筛选[J]. 中国水产科学, 2011, 18(4): 801-808. https://cstj.cqvip.com/Qikan/Article/Detail?id=38618368&from=Qikan_Search_Index [30] 李渝成, 李康, 周暾. 十四种淡水鱼的DNA含量[J]. 遗传学报, 1983, 10(5): 384-389. -
期刊类型引用(3)
1. 何福玲, 凌正宝, 肖俊, 郭忠宝, 钟欢, 杨弘, 陈文治, 唐瞻杨, 单丹, 梁军能, 甘西, 罗永巨. 两个人工选择奥利亚罗非鱼群体系统发育及其遗传多样性分析. 南方农业学报. 2017(02): 341-349 .  百度学术
百度学术
2. 颉晓勇, 钟金香, 赵金良. 遗传标记在罗非鱼良种选育研究中的应用与展望. 湖南农业科学. 2017(02): 123-126 .  百度学术
百度学术
3. 颉晓勇, 钟金香, 李思发. 3种分子标记分析罗非鱼选育群体的多态性效率比较. 广东农业科学. 2014(24): 114-117 .  百度学术
百度学术
其他类型引用(2)



 下载:
下载:
 粤公网安备 44010502001741号
粤公网安备 44010502001741号
