Proteomic analysis of granule of hemocytes from Mytilus coruscus
-
摘要:
贝类的血细胞在宿主的免疫防御机制中发挥着重要的作用。文章利用线性离子阱-四级杆质谱对厚壳贻贝(Mytilus coruscus)血细胞的颗粒蛋白质组分进行了Shotgun分析,结合Mascot数据库搜索,共鉴定了125种高丰度蛋白,其中包括细胞骨架蛋白、细胞粘附蛋白、精氨酸激酶和代谢相关酶类等。此结果为深入了解厚壳贻贝血细胞颗粒的蛋白组成、贻贝免疫分子标记的筛选及其免疫机制奠定了基础。
-
关键词:
- 厚壳贻贝 /
- 血细胞颗粒 /
- LTQ-Orbitrap /
- 质谱 /
- Shotgun
Abstract:The blood cells of mussel play an important role in the host′s immune defense mechanism. We explore the protein composition in the granule of hemocytes from Mytilus coruscus by using linear-ion-trap-quadrupole mass spectrometry (LTQ-Orbitrap) combined with Shotgun strategy. A total of 125 high-abundance proteins are identified by Mascot searching methods, including cytoskeletal proteins, cell adhesion proteins, arginine kinases and metabolism-related enzymes, etc. The results lay a foundation for further studies on the composition of mussel protein particles, screening of its immune molecular markers as well as its immune mechanism.
-
Keywords:
- Mytilus coruscus /
- hemocytes granule /
- LTQ-Orbitrap /
- mass spectrometry /
- Shotgun
-
卵形鲳鲹(Trachinotus ovatus)俗称金鲳、黄腊鲳、红三等,隶属鲈形目、鲈亚目、鲹科、鲳鲹亚科、鲳鲹属,广泛分布于太平洋、大西洋和印度洋热带及温带海域[1-3]。该鱼体型较大、生长快、食性简单、肉质细嫩鲜美,广受养殖户和消费者喜爱。随着其繁育技术获得突破,且养殖过程中可全程使用人工配合饲料,出口加工市场大等;近年来,卵形鲳鲹养殖产业在我国发展迅速,已成为广东、广西、海南等华南沿海地区网箱主养海水鱼类之一,2017年全国养殖产量超10×104 t[4-6]。
开展鱼类摄食节律研究,通过调整鱼类的摄食活动时间,使其摄食节律与大多数体细胞的生长增殖周期同步,是一种重要的时间生物学策略[7-11]。目前已有多种鱼类的摄食节律被研究,如大鳞副泥鳅(Paramisgurnus dabryanus)[12]、高体革䱨(Scortum barcoo)[13]、牙鲆(Paralichthys olivaceus)[14]、鬼鲉(Inimicus japonicus)[11]、双棘黄姑鱼(Nibea diacanthus)[15]、大口胭脂鱼(Ictiobus Cyprinellus)[16]和塞内加尔鳎(Solea senegalensis)[17]等,但对卵形鲳鲹的相关研究尚未见报道。
鱼类的耗氧率是有氧代谢强度的重要指标之一,耗氧率高低通常与鱼类的大小和摄食水平相对应,鱼类摄食活动的旺盛期通常也是高耗氧率时期。耗氧节律能直接反映出鱼类的新陈代谢规律和生理反应[18-20],耗氧率低时鱼多处在静止或缓慢运动状态,能量代谢强度低;耗氧率高时多处在剧烈运动状态,能量代谢强度高。胃肠排空是指食物从鱼摄食后至分别经胃、肠消化后排出体外的过程,胃肠排空影响着鱼类的食欲,鱼类胃肠饱满度和胃肠排空速率决定摄食量[21-23]。研究表明,不同鱼类的摄食、耗氧节律和胃肠排空时间存在较大差异。本研究以卵形鲳鲹为实验对象,探究其摄食、耗氧节律和胃肠排空时间的规律性,为确定卵形鲳鲹适宜投喂时间提供理论依据,也为其他水产动物的摄食节律研究和投喂策略提供参考。
1. 材料与方法
1.1 实验材料
本研究实验鱼为深圳大鹏澳海域中国水产科学研究院南海水产研究所深圳试验基地抗风浪网箱养殖的4月龄卵形鲳鲹,2018年9月30日,将实验鱼在露天高位池塘过渡驯养15 d,实验开始前1周,挑选大小一致、活力好且体表无伤的个体暂养于室内养殖桶中,使实验鱼适应实验环境,能在实验条件下正常摄食。养殖和暂养全程使用广东越群海洋生物研究开发有限公司生产的浮性卵形鲳鲹专用配合饲料。实验时挑选60尾用于摄食节律实验,9尾用于耗氧节律实验,90尾用于胃肠排空实验。实验期间水温为25.1~26.3 ℃、盐度32.5、pH 7.6~8.5。
1.2 实验方法
摄食节律实验采用分段式连续投喂法[10, 12],实验于2018年10月22日,将一昼夜分为12个时间段(00:00、02:00、04:00、06:00、08:00、10:00、12:00、14:00、16:00、18:00、20:00、22:00),每时段连续投喂,实验设3个平行组,每组20尾实验鱼,总质量(1 532.57±20.64) g,养殖密度约为40尾·m−3。分组后实验鱼停料1 d,从10月23日14:00开始投喂,为尽量排除实验鱼补偿摄食的影响,前3次投喂数据舍去。每次投喂前准确称取过量饲料,确保达到表观饱食,1 h后将残余饵料捞出,收集到各平行组各时段对应的小盒中,−20 ℃冰柜保存,待实验完成后,经60 ℃烘干获得实验鱼实际摄食量。测定残饵的溶失率以校正实验鱼的摄食量,即在实验完成后,将实验鱼捞出,向每组养殖桶中投入10 g实验用饲料,1 h后收集,与10 g未浸泡饲料同时放60 ℃烘干并称质量,求得溶解掉的饲料质量,计算溶失率。实验周期为3.5 d。
耗氧节律实验采用流水呼吸法[18,20],随机挑9尾卵形鲳鲹,设3个平行组,每组3尾,总质量(227.49±6.04) g,分别放入自制透明玻璃鱼缸中,用封口膜密封,玻璃缸规格为长×宽×高=40 cm×40 cm×50 cm,养殖密度约为38尾·m−3。提前用1 t水体的养殖桶备满沙滤自然海水,确保桶中水溶氧大于6 mg·L−1,调整流速,使流出实验玻璃鱼缸的水溶氧大于4 mg·L−1,实验持续1昼夜,分12次取样读取溶氧数据,取样时间与摄食节律实验相同(00:00、02:00、04:00、06:00、08:00、10:00、12:00、14:00、16:00、18:00、20:00、22:00),溶解氧用校准后的德国WTW多参数分析仪测定。
胃肠排空实验采用一次饱食投喂法[10,22],将一昼夜等分为12个时间段,每隔2 h取样1次作为一个处理组,设3个平行组,每组30尾,总质量(2 295.6±48.26) g,养殖密度约为60尾·m−3。分组后实验鱼停料2 d,确保实验鱼胃肠内容物充分排空,实验桶内无其他可食用物,投入的饲料为唯一食物来源。实验鱼饱食投喂后捞出剩饵,分别在饱食后第1、第3、第5、第7、第9、第11、第13、第15、第17、第19、第21和第23小时随机捞出2尾进行解剖取样。解剖前先用丁香酚将实验鱼深度麻醉,以减少胃肠蠕动而造成的实验误差。分别取出胃和肠中内含物并收集到各平行组各时段对应的小盒中,−20 ℃冰柜保存,待实验完成后,经60 ℃烘干称质量,实验均持续1昼夜。
1.3 数据分析
$$ \begin{array}{*{20}{c}} {{K_{\rm{p}}} = {W_{\rm{f}}} \times {W_0}^{ - 1} \times 100\% }\\ {{K_{\rm{a}}} = {W_{\rm{f}}} \times {W_0}^{ - 1} \times {D^{ - 1}} \times 100\% }\\ {{R_{\rm{s}}} = {W_{\rm{s}}} \times {W_{\rm{w}}}^{ - 1} \times 100\% }\\ {{R_{\rm{b}}} = {W_{\rm{b}}} \times {W_{\rm{w}}}^{ - 1} \times 100\% }\\ {{R_{{\rm{OCR}}}} = A \times V \times {W_{\rm{w}}}^{ - 1}\times {t^{ - 1}} \times 100\% } \end{array} $$ 式中Kp为日摄食率;Ka为平均摄食率;Rs为胃内含物比率;Rb为肠内含物比率;ROCR为耗氧率;Wf为摄食量(g);W0为初始体质量(g);D为实验天数(d);Ws为胃内含物干质量(g);Wb为肠内含物干质量(g);Ww为鱼体湿质量(g);A为消耗水溶解氧质量浓度(mg·L−1);V为容积(L);t为呼吸时间(h)。
所有统计分析由Excel 2016和SPSS 20.0软件完成,实验数据先做方差齐性检验,然后进行单因素方差分析(One-Way ANOVA),再对不同处理组进行Duncan's多重比较,取P<0.05为差异显著。
2. 结果
2.1 昼夜摄食节律
在一昼夜的分段式连续投喂下,卵形鲳鲹在上午10:00和下午14:00—16:00期间表现出2个摄食高峰(P<0.05),凌晨02:00—08:00摄食水平显著低于其他时段(P<0.05),06:00摄食水平最低(P<0.05),属于典型的白天摄食类型。第1天的摄食最高峰出现在晚上22:00,之后在上午10:00和下午16:00各出现1个摄食高峰。第2天的摄食率波动较小,凌晨06:00天亮后摄食率逐渐升高,在傍晚18:00出现1个摄食高峰。第3天的摄食节律比前2 d相对稳定,分别在晚上20:00和上午10:00出现2个摄食高峰(图1)。
![]() 图 1 在分段式连续投喂下卵形鲳鲹的昼夜摄食节律变化 (
图 1 在分段式连续投喂下卵形鲳鲹的昼夜摄食节律变化 ($ \overline X \pm {\rm{SE}}$ ,n=3)图中不同英文字母表示每个取样时间点所得数据之间达到显著差异水平(P<0.05);后图同此Fig. 1 Variation of diet feeding rhythm of T. ovatus by a continuous feeding at a fixed intervalThe values with different lowercase letters are significantly different at P<0.05 at each sampling time; the same case in the following figures.实验测得饲料的溶失率为6.3%,卵形鲳鲹的昼夜平均摄食率为2.33%,算出实验中卵形鲳鲹饲料的日摄食量为鱼体体质量的2.49%。
2.2 昼夜耗氧节律
水温25.1~26.3 ℃,卵形鲳鲹昼夜耗氧率波动较大,凌晨00:00—08:00,耗氧率逐渐升高,08:00达第1个耗氧高峰,之后迅速下降,在10:00—12:00时间段耗氧率达一天的最低值[0.413 mg·(g·h)−1](P<0.05),午后又迅速上升,在16:00达到第2个耗氧高峰,也是一天耗氧率最高值[0.702 mg·(g·h)−1](P<0.05),之后逐渐下降,至00:00耗氧率接近最低水平[0.417 mg·(g·h)−1] (P<0.05,图2)。
卵形鲳鲹在白天(8:00—18:00)平均耗氧率为(0.571±0.14) mg·(g·h)−1,夜间(18:00—08:00)平均耗氧率为(0.546±0.07) mg·(g·h)−1,白天与夜间相近(P>0.05)。
2.3 胃肠排空时间
卵形鲳鲹摄食后24 h内胃和全肠排空时间见图3。饱食后胃内含物比率下降迅速(P<0.05),7 h下降近50%,第15小时出现极低值(P<0.05),19 h后胃内含物为0。摄食后胃内含物比率呈阶梯下降趋势明显,通过与摄食后时间拟合后获得公式y=0.018 5x2−0.394x+2.071 5 (R2=0.995 5),y为胃内含物比率,x为摄食后时间。
饱食后3 h内全肠内含物比率迅速升高,第9~第11小时达到最大值(P<0.05),之后逐渐降低,在第21小时出现极低值(P<0.05)。
3. 讨论
3.1 昼夜变化对摄食率的影响
Helfman[24]把鱼类的摄食归纳为白天摄食、晚上摄食、晨昏摄食和无明显节律摄食4种类型。本研究的卵形鲳鲹第1天的摄食率波动大,第1个摄食最高峰出现在晚上22:00,之后在上午10:00和下午16:00各出现1个摄食高峰。第2天的摄食率波动较小,凌晨06:00天亮后摄食率逐渐升高,在傍晚18:00出现1个摄食高峰。第3天的摄食节律比前2天相对稳定,分别在晚上20:00和上午10:00出现2个摄食高峰。综合3 d的摄食率变化发现,其原因可能是实验鱼饥饿1 d后,补偿摄食的影响未完全消除而出现的结果,后两天卵形鲳鲹的摄食节律相对明显,表现为在上午10:00和下午14:00—16:00期间出现2个摄食高峰(P<0.05),凌晨02:00—08:00摄食水平显著低于其他时段(P<0.05),06:00摄食水平最低(P<0.05),这表明卵形鲳鲹属于典型的白天摄食类型。叉尾斗鱼(Macropodus opercularis)仔鱼在12:00—16:00表现出明显的摄食高峰,且在持续光照下摄食活动昼夜均很活跃,表明叉尾斗鱼为白天摄食类型[25]。白天摄食类型的鱼类,其视觉在摄食中具有重要意义,而夜间或晨昏摄食类型的鱼类,主要依靠如化学感觉、触觉等进行捕食[10-12, 26]。如大鳞副泥鳅[12]因长期生活在水体底层,栖息环境光线微弱,眼睛逐渐退化,触须发达,靠触觉和化学感觉来识别食物。因此按照自然摄食节律来确定适宜的喂料时间,可有效提高饲料效率,增加经济效益。可见鱼类的日摄食节律不仅能被规律的人工投喂时间适当调整,同时也受栖息环境和不同种属的影响。
3.2 昼夜变化对耗氧率的影响
鱼类的耗氧率高低通常与鱼类的大小、水温和摄食水平相对应。水温适宜,鱼类摄食活动旺盛时耗氧率也会相应升高。耗氧节律能直接反映出鱼类的新陈代谢规律和生理反应[18-20]。鱼类耗氧率的昼夜节律可分为4种,分别是白天耗氧率高于夜间耗氧率、夜间耗氧率高于白天耗氧率、昼夜耗氧率无差异和某一时间段内出现耗氧率高峰区域等[18]。本实验结果表明,卵形鲳鲹属于昼夜耗氧率基本无差异,白天和夜晚均有耗氧高峰期与低峰期,在06:00—8:00和14:00—18:00出现耗氧率高峰(P<0.05),说明卵形鲳鲹在这2个时间段呼吸代谢旺盛,活动频繁,结合前面分析的摄食节律,可能与捕食或消化有关。而在22:00—00:00和10:00—12:00出现耗氧率低谷(P<0.05)。自然条件下,22:00—00:00时间段光线微弱,对卵形鲳鲹这种白天摄食类型的鱼类,摄食活动相对减少,而10:00—12:00时间段,正处于一天中阳光直射、光照强烈且水温较高的时候,鱼类因其本性,减少活动以躲避强光。
3.3 胃肠排空时间
胃肠排空影响着鱼类的食欲,鱼类胃肠饱满度和胃肠排空速率决定摄食量[21-23]。本实验发现卵形鲳鲹在摄食7 h后约50%胃内含物已排出,第19小时胃内含物为0,全肠内含物也降至最低。余方平等[27]用相同方法测得眼斑拟石首鱼(Sciaenops ocellatus)摄食28 h后接近排空;马彩华等[28]发现大菱鲆(Scophthalmus maximus)在摄食6 h后开始排粪,20 h排空;董桂芳等[10]的研究发现斑点叉尾鮰 (Ictalurus punctatus)和杂交鲟(Acipenser baeri×A. gueldenstaedtii)胃和肠排空需要24 h。而卵形鲳鲹摄食胃肠排空时间较短,这可能与不同鱼类的胃肠消化及吸收速率有关。在同一时间段,胃内含物的减少量大于肠内含物的增加量,这可能是由于卵形鲳鲹属于游动迅速、活动量大的鱼类,为适应快速游动,身体侧扁,胃相对较小,肠道也相应较短,因此食物在体内被消化和吸收快,甚至可能部分营养物质在胃内已被消化吸收。结合昼夜节律实验发现,卵形鲳鲹在胃肠排空之前就有摄食行为,且全时段都能进行摄食。
3.4 投喂策略
投喂策略是鱼类养殖技术的重要部分,投喂不当不仅浪费饵料和劳动力,而且污染水质导致养殖效益低[26]。科学的投喂策略应符合鱼类摄食、耗氧节律和胃肠排空时间等生活习性和生理变化。研究表明鱼类摄食节律的形成与自然界食物丰度有关,推测长期特定的人工投喂策略可改变鱼类的摄食节律[29]。因此可通过特定时间的投喂来驯化鱼类的摄食时间,从而调整其日摄食节律。Luo等[30]研究了杂交鲟(A. schrenckii ♀×A. baeri ♂)育成阶段的最适投喂频率和投喂率。表明投喂频率和投喂率均对鲟鱼的生长性能指标有极显著影响(P<0.01),鲟鱼育成阶段最适投喂策略为以3.7%鱼体质量的投喂率,每天投喂6次。郑伟力等[31]通过对大鳞副泥鳅繁育研究表明可通过人工养殖驯化,将大鳞副泥鳅夜间摄食节律调整为晨昏摄食。因此,生产上可根据实际养殖情况对卵形鲳鲹进行定时定点的人工投饵驯化,适当调整其摄食节律,以提高人工养殖效率。
综合卵形鲳鲹昼夜摄食、耗氧节律和胃肠排空的实验结果,建议在卵形鲳鲹网箱养殖生产中,宜在光线较强和耗氧、摄食高峰的上午(09:00—10:00)和下午(14:00—16:00)时段进行投喂,投喂频率2~3次·d−1,投喂间隔7~9 h。
-
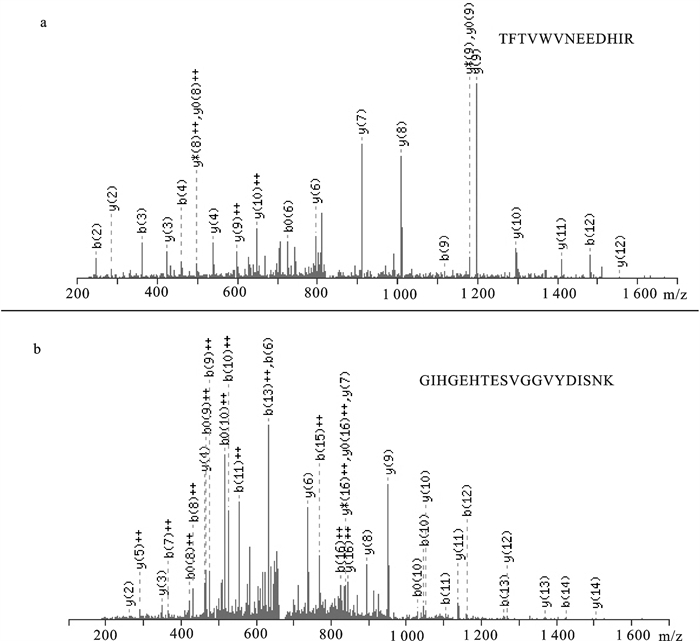
图 2 表 1中序号为3的蛋白质MS/MS图谱
a. m/z为1 656.84 Da的MS/MS图谱,分别显示了其b离子系列和y离子系列的单电荷峰及双电荷峰(“+ +”标注),由此推导的肽段氨基酸序列为“TFLVWVNEEDHIR”;b.m/z为1 997.95 Da的MS/MS图谱,分别显示了其b离子系列和y离子系列的单电荷峰及双电荷峰(“+ +”标注),由此推导的肽段氨基酸序列为“GIHGEHTESVGGVYDISNK”;其鉴定结果为精氨酸激酶(来自Liolophura japonica)
Figure 2. Tandem mass spectrum of No.3 protein in Tab. 1
a. m/z indicates MS/MS spectrum with m/z 1 656.84 acquired on LTQ-Orbitrap mass spectrometer and the amino acid sequence "TFLVWVN-EEDHIR" deduced from precise mass difference between adjacent b-and y-ions; b. m/z indicates MS/MS spectrum with m/z 1 997.95 acquired on LTQ-Orbitrap mass spectrometer and the amino acid sequence "GIHGEHTESVGGVYDISNK" deduced from precise mass difference between adjacent b-and y-ions. The results of the Mascot search are presented as arginine kinase from Liolophura japonica.
表 1 LTQ质谱MS/MS数据的搜库结果
Table 1 Protein identification of hemocyte granule from M.coruscus by LTQ-Orbitrap with Mscot searching
序号
No.数据库编号
accession No.in UniProtKB同源性
identity/homology物种
species匹配肽段
matched peptide得分
score1 B4J603_DROGR GH21113 Drosophila grimshawi 1 305 2 Q6UQ16_MYTED EP protein Mytilus edulis 3 258 3~4 KARG_LIOJA arginine kinase Liolophura japonica 2 215 5 B3MCR5_DROAN GF11480 Drosophila ananassae 1 209 6 ACT_PLAMG actin Placopecten magellanicus 6 209 7 Q5BQE3_9VEST actin A3 Haliotis iris 5 209 8~9 Q760P6_CRAGI arginine kinase Crassostrea gigas 3 179 10 B3N2H1_DROAN GF20391 Drosophila ananassae 4 153 11 Q9NC09_9MOLL actin Mastigoteuthis magna 1 137 12 ACT_MAYDE actin Mayetiola destructor 4 111 13~14 D7GXP9_TRICA histone H2B Tribolium castaneum 3 111 15 Q17C86_AEDAE actin Aedes aegypti 4 103 16 Q2LDZ7_HIRME cytoplasmic actin Hirudo medicinalis 3 98 17 MLE_TODPA myosin catalytic light chain Todarodes pacificus 1 97 18 Q9NC07_9MOLL actin (fragment) Alluroteuthis antarcticus 3 87 19 D0ES27_9HYME beta actin Polyrhachis vicina 6 84 20~24 B3VN78_BOMMOL.L-1A actin Bombyx mandarina 5 84 25 B4LVN2_DROVI histone H4 Drosophila virilis 1 79 26 B2YGD6_9ARAC actin Goleba lyra 2 79 27~30 B0FRF9_LITVA arginine kinase Litopenaeus vannamei 1 78 31 B4LB51_DROVI GJ12598 Drosophila virilis 1 78 32 B6EAU5_HOMAM skeletal muscle actin Homarus americanus 4 78 33 B3MQE5_DROAN histone H2A Drosophila ananassae 2 77 34 A4FT58_9BILA actin Macrobiotus sp. 3 76 35~37 B4KDT0_DROMO histone H2A Drosophila mojavensis 2 75 38 C7SP19_9BILA fructose-bisphosphate aldolase Glottidia sp. 1 75 39~41 E2IV58_HELAM actin Helicoverpa armigera 4 72 42 Q7Q944_ANOGA AGAP004835-PA Anopheles gambiae 2 70 43~45 B2YGB1_9ARAC actin Habronattus americanus 4 70 46 B0LUD9_LYMDI beta-actin Lymantria dispar 2 70 47 TPM_MYTED tropomyosin Mytilus edulis 2 66 48~50 Q966R4_9DIPT actin Chironomus yoshimatsui 2 64 51 D2A3B9_TRICA putative protein Tribolium castaneum 1 62 52 B0X6X2_CULQU dumpy Culex quinquefasciatus 1 60 53 B3MD57_DROAN GF11439 Drosophila ananassae 1 59 54 B4JTF2_DROGR GH24039 Drosophila grimshawi 1 59 55 B4N5S6_DROWI GK17871 Drosophila willistoni 1 59 56 Q6S015_DROME reverse transcriptase Drosophila melanogaster 1 59 57 Q7PYN5_ANOGA AGAP002033-PA Anopheles gambiae 1 59 58 Q6X4W3_HAELO actin Haemaphysalis longicornis 5 59 59 D2DGZ3_9CUCU cytoplasmic actin Ips confusus 2 59 60 B3MDN4_DROAN GF11378 Drosophila ananassae 1 57 61 B4NFT1_DROWI GK22698 Drosophila willistoni 2 56 62 D6X1R9_TRICA putative protein Tribolium castaneum 1 56 63 Q9VLL1_DROME D12 Drosophila melanogaster 1 55 64 B4KI41_DROMO GI10553 Drosophila mojavensis 1 54 65 B0WCT3_CULQU leucine-rich transmembrane protein Culex quinquefasciatus 1 54 66 E0AD92_BOOMI angiotensin-converting enzyme-like protein Boophilus microplus 1 50 67 E2B904_9HYME putative protein Harpegnathos saltator 1 50 68 D6WDM1_TRICA putative protein Tribolium castaneum 2 49 69 B0W6D8_CULQU survivin Culex quinquefasciatus 2 49 70 A2AXC2_TRICA gustatory receptor Tribolium castaneum 1 49 71 B0XCW8_CULQU putative protein Culex quinquefasciatus 1 49 72 B4MJJ2_DROWI GK20842 Drosophila willistoni 1 48 73 B3P4Z6_DROER GG11834 Drosophila erecta 1 48 74 E2BDR6_9HYME lipid storage droplets surface-binding protein 1 Harpegnathos saltator 1 48 75 B7UEY4_MYTGA superoxide dismutase Mytilus galloprovincialis 1 47 76 B4NIN2_DROWI GK13512 Drosophila willistoni 1 47 77 B0XJN8_CULQU phosphatase Slingshot Culex quinquefasciatus 1 47 78 E2BKC0_9HYME MAP kinase Harpegnathos saltator 1 47 79 E2BI22_9HYME N-acetylglucosamine-6-sulfatase Harpegnathos saltator 1 46 80 Q75W49_CRAGI 78kDa glucose regulated protein Crassostrea gigas 2 46 81 D7EKZ5_TRICA putative protein Tribolium castaneum 1 46 82 E2AMK0_9HYME DE-cadherin Camponotus floridanus 1 46 83 B0W9Q9_CULQU putative protein Culex quinquefasciatus 1 46 84 B3MMOL.L-1U1_DROAN GF15119 Drosophila ananassae 1 45 85 Q95UF1_DROSI ankyrin Drosophila simulans 1 45 86 B4KUQ4_DROMO GI11592 Drosophila mojavensis 1 45 87 Q16WC1_AEDAE putative protein Aedes aegypti 1 45 88 Q1HPK0_BOMMOL.L-1O vesicle amine transport protein Bombyx mori 1 45 89 D6X0H5_TRICA putative protein Tribolium castaneum 1 45 90 E0VRW9_PEDHC putative protein Pediculus humanus 1 45 91 B4M706_DROVI GJ16541 Drosophila virilis 2 45 92 B0W417_CULQU ataxia telangiectasia Culex quinquefasciatus 1 45 93 Q7PK24_ANOGA AGAP009815-PA Anopheles gambiae 1 45 94 D0IQG5_DROME MIP13274p Drosophila melanogaster 1 44 95 B7P8Q8_IXOSC putative protein Ixodes scapularis 1 44 96 B3ME79_DROAN GF12460 Drosophila ananassae 1 44 97 D7F165_BOMMOL.L-1O endonuclease-reverse transcriptase Bombyx mori 1 44 98 D7EKS8_TRICA putative protein Tribolium castaneum 1 43 99 D7ELX7_TRICA putative protein Tribolium castaneum 1 43 100 Q9U0S5_MYTGA catchin protein Mytilus galloprovincialis 2 43 101 Q9U0S7_MYTGA myosin heavy chain Mytilus galloprovincialis 1 43 102 B4JUI7_DROGR GH15720 Drosophila grimshawi 1 43 103 Q966V3_MYTGA calponin-like protein Mytilus galloprovincialis 5 43 104 Q16SH8_AEDAE putative protein Aedes aegypti 1 43 105 E2BDB9_9HYME putative protein Harpegnathos saltator 1 43 106 MLE_AEQIR myosin essential light chain Aequipecten irradians 1 43 107 E2C3M9_9HYME hexokinase-2 Harpegnathos saltator 1 43 108 B5DNP1_DROPS GA22337 Drosophila pseudoobscura 1 42 109 A7UTW9_ANOGA AGAP005832-PA Anopheles gambiae 1 42 110 B7P2A1_IXOSC 26S proteasome regulatory complex Ixodes scapularis 1 42 111 B0WTT0_CULQU putative protein Culex quinquefasciatus 1 42 112 B7PXU8_IXOSC CAP-Gly linker protein Ixodes scapularis 1 42 113 B0WAH4_CULQU WD repeat protein 36 Culex quinquefasciatus 1 42 114 E2A5M0_9HYME putative protein Camponotus floridanus 1 42 115 B4JPX7_DROGR GH13313 Drosophila grimshawi 1 42 116 C4WX22_ACYPI ACYPI004711 protein Acyrthosiphon pisum 1 42 117 B4J2K3_DROGR GH16634 Drosophila grimshawi 1 42 118 Q0C728_AEDAE golgi protein Aedes aegypti 1 42 119 E0VRY6_PEDHC guanine nucleotide exchange factor DBS Pediculus humanus subsp. corporis 1 41 120 D2T1I1_9NEOP cadherin-like protein Polyplectropus sp. 1 41 121 Q16U82_AEDAE putative protein Aedes aegypti 1 41 122 E0VM19_PEDHC alpha-actinin-4 Pediculus humanus 1 41 123 D1M9K5_9MAXI elongation factor 1 Notochthamalus scabrosus 1 40 124 E2A3P3_9HYME N-acetylgalactosaminyltransferase Camponotus floridanus 1 40 125 E2C7U1_9HYME putative protein Harpegnathos saltator 1 40 注:按数据库得分从高到低排列,展示得分在40分以上的结果
Note: Scores are ranked from high to low,and those more than 40 are shown. -
[1] SUN Jingfeng, WU Xinzhong. Histology, ultrastructure and morphogenesis of a Rickettsi-like organism causing disease in the oyster, Crassostrea ariakensis [J]. Invertebr Pathol, 2004, 86(3): 77-86. doi: 10.1016/j.jip.2004.04.004
[2] CHARLET M, CHERNYSH S, PHILIPPE H, et al. Isolation of several cysteine-rich antimicrobial peptides from the blood of a mollusk, Mytilus edulis [J]. J Biol Chem, 1996, 271(36): 21808-21813. doi: 10.1074/jbc.271.36.21808
[3] MITTA G, VANDENBULCKE F, HUBERT F, et al. Involvement of Mytilins in mussel antimicrobial defense[J]. J Biol Chem, 2000, 275(17): 12954-12962. doi: 10.1074/jbc.275.17.12954
[4] YANG Y S, MITTA G, CHAVANIEU A. Solution structure and activity of the synthetic four-disulfide bond Mediterranean mussel defensin (MGD-1)[J]. Biochemistry, 2000, 39(47): 14436-14447. doi: 10.1021/bi0011835
[5] MITTA G, HUBERT F, DYRYNDA E A, et al. Mytilin B and MGD-2, two antimicrobial peptides of marine mussels: gene structure and expression analysis[J]. Dev Comp Immunol, 2000, 24(4): 381-393. doi: 10.1016/S0145-305X(99)00084-1
[6] MITTA G, HUBERT F, NOEL T, et al. Myticin, a novel cysteine-rich antimicrobial peptide isolated from hemocytes and plasma of the mussel Mytilus galloprovincialis [J]. Eur J Biochem, 1999, 265(1): 71-78. doi: 10.1046/j.1432-1327.1999.00654.x
[7] FRIEBEL B, RENWRANTZ L. Application of density gradient centrifugation for separation of eosinophilic and basophilic haemocytes from Mytilus edulis and characterization of both cell groups[J]. Comp Biochem Physiol, 1996, 112(1): 81-90. doi: 10.1016/0300-9629(95)00086-M
[8] BAYNE C J, MOORE M N, CAREFOOT T H, et al. Hemolymph functions in Mytilus californianus: the cytochemistry of hemocytes and their responses to foreign implants and hemolymph factors in phagocytosis[J]. Invertebr Pathol, 1979, 34(1): 1-20. doi: 10.1016/0022-2011(79)90048-X
[9] CHENG T C. Bivalves invertebrate blood cells[M]. New York: Academic Press, 1981: 233-330. https://xueshu.baidu.com/usercenter/paper/show?paperid=b14f0fcedf73f9686489101264690ac9&site=xueshu_se
[10] MOORE M N, LOWE D M. The cytology and cytochemistry of the hemocytes of Mytilus edulis and their response to experimentally injected carbon particles[J]. Invertebr Pathol, 1977, 29(1): 18-30. doi: 10.1016/0022-2011(77)90167-7
[11] 王日昕, 廖智, 刘梅, 等. 厚壳贻贝血细胞cDNA文库的构建及部分EST序列分析[J]. 海洋与湖沼, 2009, 40(5): 603-607. doi: 10.3321/j.issn:0029-814X.2009.05.013 WANG Rixin, LIAO Zhi, LIU Mei, et al. Construction of haemolymphe cDNA library of Mytilus coruscus and the partial screening of expressed sequence tags[J]. Oceanologia et Limnologia Sinica, 2009, 40(5): 603-607. (in Chinese). doi: 10.3321/j.issn:0029-814X.2009.05.013
[12] 廖智, 刘梅, 王日昕, 等. 厚壳贻贝抗菌肽Mytilin和Myticin的cDNA基因的克隆与序列分析[J]. 水产学报, 2010, 34(7): 1025-1033. doi: 10.3724/SP.J.1231.2010.06878 LIAO Zhi, LIU Mei, WANG Rixin, et al. cDNA clone and sequence analysis of mytilin and myticin from Mytilus coruscus[J]. J Fish China, 2010, 34(7): 1025-1033. (in Chinese). doi: 10.3724/SP.J.1231.2010.06878
[13] 王日昕, 刘梅, 廖智, 等. 厚壳贻贝抗菌肽Mytilin的初步鉴定[J]. 水产学报, 2010, 334(1): 153-159. doi: 10.3724/SP.J.1231.2010.06514 WANG Rixin, LIU Mei, LIAO Zhi, et al. Purification and identification of Mytilins from Mytilus coruscus [J]. J Fish China, 2010, 334(1): 153-159. (in Chinese). doi: 10.3724/SP.J.1231.2010.06514
[14] BACHÉRE E, HERVIO D, MIALHE E, et al. Luminol-dependent chemiluminescence by hemocytes of two marine bivalves, Ostrea edulis and Crassostrea gigas [J]. Dis Aquat Org, 1991, 11: 173-180. doi: 10.3354/dao011173
[15] LEIPPE M, RENWRANTZ L. Release of cytotoxic and agglutinating molecules by Mytilus hemocytes[J]. Dev Comp Immunol, 1988, 12(2): 297-308. doi: 10.1016/0145-305X(88)90006-7
[16] ROCH P. Defense mechanisms and disease prevention in farmed marine invertebrates[J]. Aquaculture, 1999, 172(1/2): 125-145. doi: 10.1016/S0044-8486(98)00439-6
[17] BACHERE E, MIALHE E, NOEL D, et al. Knowledge and research prospects in marine mollusk and crustacean immunology[J]. Aquaculture, 1995, 132(1/2): 17-32. doi: 10.1016/0044-8486(94)00389-6
[18] PIPE R K. Hydrolytic enzymes associated with the granular haemocytes of the marine mussel Mytilus edulis [J]. Histochem J, 1990, 22(11): 595-603. doi: 10.1007/BF01072941
[19] PAN C, PARK B H, MCDONALD W H, et al. A high-thoughput de novo sequencing approach for Shotgun proteomics using high-resolution tandem mass spectrometry[J]. BMC Bioinformatics, 2010, 11(1): 118. doi: 10.1186/1471-2105-11-118
[20] HERBINIERE, PIERRE G, JEAN-MARC S, et al.Protein profiling of hemocytes from the terrestrial crustacean Armadillidium vulgare [J]. Dev Comp Immunol, 2008, 32(8): 875-882. doi: 10.1016/j.dci.2008.01.007
[21] 贺淹才. 肌动蛋白和肌动蛋白基因的研究进展[J]. 生命的化学, 2002, 22(3): 248-250. doi: 10.3969/j.issn.1000-1336.2002.03.017 HE Yancai. Research progress in actin and its gene[J]. Chem Life, 2002, 22(3): 248-250. (in Chinese) doi: 10.3969/j.issn.1000-1336.2002.03.017
[22] YOSHIDA S, SASAKAWA C. Exploiting host microtubule dynamics: a new aspect of bacterial invasion[J]. Trends Microbiol, 2003, 11(3): 139-143. doi: 10.1016/s0966-842x(03)00023-4
[23] PHATTARA-ORN C, APICHAI B, CHU-FANG L, et al. Proteomic analysis of differentially expressed proteins in Penaeus vannamei hemocytes upon Taura syndrome virus infection[J]. Proteomics, 2007, 7(19): 3592-3601. doi: 10.1002/pmic.200700281
[24] YAO Cuiluan, WU Changgong, XIANG Jianhai, et al. Molecular cloning and response to laminarin stimulation of arginine kinase in haemolymph in Chinese shimp, Fenneropenaeus chinensis[J]. Fish Shellfish Immunol, 2005, 19(4): 317-329. doi: 10.1016/j.fsi.2005.01.006
[25] ASTROFSKY K M, ROUX M M, KLIMPEL K R, et al. Isolation of differentially expressed genes from white spot virus (WSV) infected Pacific blue shimp (Penaeus stylirostris)[J]. Arch Virol, 2002, 147(9): 1799-1812. doi: 10.1007/s00705-002-0845-z
[26] NAKAMURA M, MORI K, INOOKA S, et al. In vitro production of hydrogen peroxide by the hamoebocytes of the scallop, Potinopecten yessoensis[J]. Dev Comp Immunol, 1985, 9(3): 407-417. doi: 10.1016/0145-305x(85)90004-7
[27] PIPE R K. Generation of reactive oxygen metabolites by the haemocytes of the mussel Mytilus edulis[J]. Dev Comp Immunol, 1992, 16(2/3): 111-112. doi: 10.1016/0145-305X(92)90012-2
[28] ROSERBERGER C M, FINLAY B B. Phagocyte sabotage: disruption of macrophage signalling by bacterial pathogens[J]. Mol Cell Biol, 2003, 4(5): 385-396. doi: 10.1038/nrm1104
-
期刊类型引用(6)
1. 张静,戴佳玥,来新昊,刘旭祥,张浩,王学锋,汤保贵. 卵形鲳鲹应对流速胁迫的代谢组学分析. 海洋学报. 2023(05): 53-63 .  百度学术
百度学术
2. 段鹏飞,田永胜,李振通,李子奇,陈帅,黎琳琳,王心怡,王林娜,刘阳,李文升,王晓梅,李波. 棕点石斑鱼(♀)×蓝身大斑石斑鱼(♂)杂交后代与棕点石斑鱼低氧耐受能力初步研究. 中国水产科学. 2022(02): 220-233 .  百度学术
百度学术
3. 逯云召,于燕光,薄其康,马超,宓慧菁,孙晓旺. 大泷六线鱼幼鱼的摄食节律研究. 渔业现代化. 2021(02): 35-39 .  百度学术
百度学术
4. 李志成,江飚,钟志鸿,李诗钰,何润真,唐嘉嘉,李安兴. 硫酸铜治疗卵形鲳鲹淀粉卵涡鞭虫病的研究. 南方水产科学. 2021(03): 108-114 .  本站查看
本站查看
5. 韩明洋,周胜杰,杨蕊,胡静,马振华. 温度胁迫下卵形鲳鲹仔鱼骨骼组织病理及分子表征. 南方农业学报. 2021(11): 3147-3156 .  百度学术
百度学术
6. 黄小林,戴超,虞为,杨洁,杨育凯,李涛,林黑着,黄忠,孙莘溢,舒琥. 丁香酚对卵形鲳鲹幼鱼的麻醉效果. 广东海洋大学学报. 2020(04): 124-131 .  百度学术
百度学术
其他类型引用(3)






 下载:
下载:


 粤公网安备 44010502001741号
粤公网安备 44010502001741号
