Optimization of SRAP-PCR system for Portunus trituberculatus based on orthogonal experimental design
-
摘要:
相关序列扩增多态性(sequence related amplified polymorphism,SRAP)是一种多态性高的新型分子标记,其扩增结果稳定,容易操作和引物通用性高,在遗传多样性分析、种质鉴定和遗传图谱构建等方面的广泛应用逐渐从植物转移到水产动物中。为了建立三疣梭子蟹(Portunus trituberculatus)SRAP技术体系,文章利用正交设计L16 (45)对三疣梭子蟹SRAP-PCR反应体系的5因素[TaqDNA聚合酶、镁离子(Mg2+)、模板、三磷酸脱氧核苷(dNTPs)和引物]在4个水平上进行优化试验,结果得出各因素水平变化对PCR反应的影响从大到小依次为模板(纯度1.75~1.90,质量浓度10~70 ng · μL-1),Mg2+(1.60~2.00 mmol · L-1),dNTPs(0.10~0.40 mmol · L-1),TaqDNA聚合酶(0.50~2.00 U)和引物(0.10~0.40 μmol · L-1);筛选出各反应因素的最佳水平,建立三疣梭子蟹SRAP-PCR反应的最佳体系(25 μL)为TaqDNA聚合酶0.50 U,Mg2+ 2.20 mmol · L-1,模板DNA 10 ng,dNTPs 0.20 mmol · L-1,引物0.20 μmol · L-1。这一优化体系可望在三疣梭子蟹遗传多样性和性别连锁标记等研究中应用。
Abstract:SRAP is a novel molecular marker with high polymorphism. It is reliable in amplification, easy to operate and has a high universality in primer pairs. SRAP technique has been widely used for studies from plants to aquatic animals in the fields of genetic diversity, germplasm identification, genetic map construction and so on. Based on an orthogonal experimental design, we optimized the five factors of SRAP-PCR amplification system for Portunus trituberculatus (TaqDNA polymerase, Mg2+, template, dNTPs and primer) from four levels. The results show that the effects of these factors on PCR reaction from strong to weak is as follows: template (purity 1.75~1.90, concentration 10~70 ng · μL-1), Mg2+ (1.60~2.00 mmol · L-1), dNTPs (0.10~0.40 mmol · L-1), TaqDNA polymerase (0.50~2.00 U) and primer (0.10~0.40 μmol · L-1).The optimal SRAP-PCR reaction system (25 μL) can be summarized as follows: 0.50 U TaqDNA polymerase, 2.20 mmol · L-1 Mg2+, 10 ng DNA template, 0.20 mmol · L-1 dNTPs and 0.20 μmol · L-1 primer. This optimized system may be helpful in the studies of genetic diversity and sex-linked markers for P.trituberculatus.
-
Keywords:
- Portunus trituberculatus /
- SRAP /
- orthogonal design /
- optimization
-
长鳍金枪鱼(Thunnus alalunga)是南太平洋延绳钓金枪鱼渔业的重要目标种类,也是中国金枪鱼延绳钓渔船的主捕对象之一[1]。联合国粮农组织(FAO)2010年统计数据显示,南太平洋海域长鳍金枪鱼产量为15.29×104 t,占当年全球长鳍金枪鱼总产量的63%,中国渔船于1998年开始进入该海域进行长鳍金枪鱼延绳钓作业,近年来其产量和作业规模总体呈现出波动增长的趋势[2-3]。相关资源评估报告表明,目前南太平洋长鳍金枪鱼资源状态总体较好,仍具有一定的开发潜力[4]。远洋性鱼类,尤其是金枪鱼,为捕食深海散射层(deep scattering layer,DSL)生物,都具有“昼潜夜浮”的特性[5]。ZAGAGLIA等[6]认为这种垂直方向上的昼夜移动,会使得金枪鱼垂直活动水层与次表层水温的关系更为密切。DOMOKOS等[7]通过在美属萨摩亚群岛附近海域进行成年长鳍金枪鱼标志放流研究发现,长鳍金枪鱼在18~30 ℃水层均有分布,其中65%~70%的时间在20~25 ℃水层活动。延绳钓调查也得出相似结论,林显鹏等[8]在南太平洋所罗门群岛以及杨嘉樑等[9]在东太平洋库克群岛附近海域进行延绳钓调查得出,长鳍金枪鱼适宜栖息水温分别为20~26 ℃以及20~25 ℃。上述研究结果表明,水温垂直结构对长鳍金枪鱼垂直活动水层的分布有直接影响,同时也是长鳍金枪鱼渔场形成过程中极为重要的因素之一。因此,分析南太平洋长鳍金枪鱼的垂直活动水层分布特征,了解其适宜的垂直活动水层深度范围十分有必要。以往对长鳍金枪鱼垂直活动水层的研究主要依靠标志放流和延绳钓调查,但标志放流和延绳钓调查的区域跨度小且在时间上不连续,无法大面积、长时间序列的监测海洋次表层环境变化对长鳍金枪鱼资源分布的影响。该研究根据前人标志放流和延绳钓调查所得结论,采用Argo数据重构了南太平洋次表层20 ℃和25 ℃等温线深度场,结合2010年~2012年中水集团长鳍金枪鱼延绳钓生产统计数据,分析了南太平洋长鳍金枪鱼的垂直分布特征,并采用数值方法计算其适宜的垂直活动水层深度,以期为南太平洋延绳钓长鳍金枪鱼捕捞作业、资源保护和管理提供参考。
1. 材料与方法
1.1 数据
1.1.1 渔业数据
渔业数据来源于中水集团南太平洋长鳍金枪鱼延绳钓船队的21艘延绳钓渔船渔捞日志,数据包括作业时间、作业位置、渔获量、渔获尾数、下钩数等信息,时间为2010年~2012年,空间范围为5°S~21°S、155°E~180°E,具体研究区域见图 1。在实际分析统计中,以1°×1°为空间统计单元,并按月计算各1°×1°渔区网格内的单位捕捞努力量渔获量(catch per unit effort,CPUE)。
CPUE计算公式如下[10]:
$$ \operatorname{CPUE}_{(i, j)}=\frac{N_{f is h(i, j)} \times 1\;000}{N_{h o o k(i, j)}} $$ (1) 式中CPUE(i, j)、Nfish(i, j)、Nhook(i, j)分别是第i个经度、第j个纬度所处渔区网格内的CPUE(尾·千钩-1)、渔获尾数和实际下钩数。随后计算2010年~2012年各月各渔区网格内所有CPUE值的四分位数(Q1~Q3),并将所有大于Q3(第3个四分位点)的CPUE称为高值CPUE,其所属渔区定义为南太平洋长鳍金枪鱼的中心渔场[11]。2010年~2012年按月统计的1°×1°渔区网格内CPUE样本数共1 939个,其中高值CPUE样本485个,CPUE的四分位数划分如下:Q1=7.27尾·千钩-1,Q2=11.23尾·千钩-1,Q3=15.78尾·千钩-1。
1.1.2 Argo数据
综合前人标志放流和延绳钓调查所得结论,选取20 ℃和25 ℃分别作为南太平洋长鳍金枪鱼适宜栖息水温的下界和上界,用于分析其垂直活动水层分布特征及适宜的垂直活动水层深度范围。2010年~2012年南太平洋次表层20 ℃和25 ℃等温线深度数据来源于中国Argo实时数据中心(http://www.argo.org.cn/)。文章采用杨胜龙等[12]的方法将离散分布的Argo剖面深度数据,等距插值到规则等温线层上,垂直等距间隔为1 m。将2010年~2012年所有Argo数据按月进行分组,分别提取20 ℃和25 ℃等温面上离散分布的深度值,并采用Kriging插值方法将其插值到1°×1°网格节点上。
1.2 方法
1.2.1 空间分析
将2010年~2012年南太平洋次表层20 ℃和25 ℃等温线深度数据与长鳍金枪鱼CPUE数据按月进行空间匹配,采用空间分析软件Arcgis绘制20 ℃和25 ℃等温线深度与CPUE空间叠加分布图。根据南半球的季节划分(1月~3月为夏季、4月~6月为秋季、7月~9月为冬季、10月~12月为春季),分析不同季节下长鳍金枪鱼CPUE与20 ℃和25 ℃等温线深度的时空分布特征及其动态变化规律。
1.2.2 垂直活动水层深度计算
采用频次分析和经验累积分布函数(empirical cumulative distribution function,ECDF)相结合的方法,计算南太平洋长鳍金枪鱼适宜的垂直活动水层深度范围[13]。1)计算南太平洋长鳍金枪鱼中心渔场20 ℃和25 ℃等温线深度的平均值、均方差及适宜的垂直活动水层深度区间(平均值±均方差);2)计算高值CPUE与20 ℃和25 ℃等温线深度的经验累积分布函数及适宜的垂直活动水层深度区间[最大D(t)处深度值±均方差];取两者交集,即为长鳍金枪鱼适宜的垂直活动水层深度。
ECDF计算方法如下[14]:
$$ f(t)=\frac{1}{n} \sum\limits_{i=1}^n l\left(x_i\right) \quad l\left(x_i\right)= \begin{cases}1, & x_i \leq t \\ 0, & x_i>t\end{cases} $$ (2) $$ g(t)=\frac{1}{n} \sum\limits_{i=1}^n \frac{y_i}{\bar{y}} $$ (3) $$ D(t)=|f(t)-g(t)| $$ (4) 式中f(t)为深度值的经验累积分布函数,g(t)为高值CPUE与深度的加权经验累积分布函数,l(t)为分段函数,D(t)为t时刻处f(t)与g(t)差的绝对值。n为高值CPUE样本个数;t是以1 m为间距从低到高排列的深度范围序列;xi为第i个样本对应的深度值;yi为第i个样本对应的CPUE;y为所有高值CPUE样本的平均值;根据给定的显著性水平α=0.05,采用双样本K-S(Kolmogorov-Smirnov)检验方法分别检验高值CPUE与20 ℃和25 ℃等温线深度间的关系。
2. 结果与分析
2.1 长鳍金枪鱼垂直活动水层空间分析
2010年~2012年南太平洋长鳍金枪鱼CPUE与20 ℃等温线深度的月平均空间分布见图 2。20 ℃等温线深度大致以15°S为中心,呈南北对称分布,且存在明显的季节性变化,长鳍金枪鱼CPUE的时空分布与20 ℃等温线深度的220 m等深线时空变动特征具有明显的相关性。1月,研究海域内20 ℃等温线深度基本在220 m以上,深度为220~230 m区域占整个研究海域面积的比例较大,高值CPUE主要分布于12°S~18°S,深度为220~240 m的海域,瓦努阿图东北部,20 ℃等温线深度小于220 m的海域,也有少量高值CPUE分布;2月~3月,研究海域南部的20 ℃等温线深度略有下降,中心渔场开始北移,渔场分布的纬度范围变窄,瓦努阿图东北部渔场消失,而深度为220~230 m的区域面积则进一步扩张,约占整个研究海域面积的一半;秋季(4月~6月),随着作业渔船数量的增加,高值CPUE的个数及分布范围均有所拓展,220 m等深线较1月~3月有明显的向南移动趋势,20 ℃等温线深度为220~230 m的海域面积开始逐步缩小,中心渔场所在区域20 ℃等温线深度则逐步加深,20 ℃等温线深度超过230 m的中心渔场数量显著增加,随着220 m等深线的南移,研究区域北部20 ℃水层的深度开始逐步变浅,而研究海域南部20 ℃等温线深度则开始逐步加深;从冬季(7月~9月)开始,10°S以北海域20 ℃等温线深度已基本全部处于220 m以下,此处区域全年均少有渔场出现,中心渔场位置开始逐步向南移动,深度为220~240 m的区域面积进一步缩小,渔场分布的纬度范围开始重新收窄;春季(10~12月),中心渔场继续向南移动,中心渔场所处海域的20 ℃等温线深度则继续加深,到12月,南太平洋长鳍金枪鱼中心渔场已完全分布于15°S以南,中心渔场所在海域的20 ℃等温线深度下界也随之拓展到250 m。
2010年~2012年南太平洋长鳍金枪鱼CPUE与25 ℃等温线深度的月平均空间分布见图 3。25 ℃等温线深度大致呈由北向南逐步递减的纬向带状分布。25 ℃等温线深度也存在明显的季节性变化,长鳍金枪鱼中心渔场的时空分布与25 ℃等温线深度的140 m等深线季节性变化趋势具有明显的相关性。1月,长鳍金枪鱼高值CPUE主要分布于25 ℃等温线深度为100~140 m的海域,瓦努阿图东北部,25 ℃等温线水深在80 m以下的海域,也有少量高值CPUE分布;2月~3月,中心渔场逐步北移,中心渔场对应的25 ℃等温线深度范围变窄,瓦努阿图东北部渔场消失;秋季(4月~6月),随着研究海域内捕捞努力量的增加,中心渔场的数量和空间分布范围均有所拓展,140 m等深线开始逐步南移,水深为140~160 m的海域面积不断扩张,中心渔场对应的25 ℃等温线深度范围拓宽至80~160 m;冬季(7月~9月),研究海域北部25 ℃等温线深度开始加深,140~160 m水深海域面积达到全年最大值,140 m等深线已完全分布于15°S以南,中心渔场对应的25 ℃等温线深度也随之变浅至80~150 m,研究海域南部25 ℃等温线深度开始变浅,中心渔场也随着140 m等深线的变动而向南转移,此处海域等深线分布密集,水层结构较为复杂,同时也是长鳍金枪鱼高值CPUE密集分布的海域;从春季(10月~12月)开始,140 m等深线开始北移,研究海域北部25 ℃等温线深度逐渐变浅,140~160 m水深的海域面积逐渐缩小,而在研究海域南部,25 ℃等温线深度则开始逐步加深,随着研究海域南部25 ℃等温线深度的增加,长鳍金枪鱼中心渔场的分布也随之不断南移,到12月,长鳍金枪鱼中心渔场已完全分布于15°S以南,中心渔场对应的25 ℃等温线深度范围缩小至80~120 m。
2.2 长鳍金枪鱼适宜垂直活动水层深度计算
2010年~2012年南太平洋长鳍金枪鱼中心渔场所在区域,20 ℃等温线深度在181~272 m,25 ℃等温线深度在41~201 m均有分布。长鳍金枪鱼高值CPUE与20 ℃和25 ℃等温线深度的频次分布直方图(图 4)呈现出明显的偏正态分布趋势,20 ℃等温线深度平均值为227 m、均方差为18,25 ℃等温线深度平均值为115 m,均方差为27。采用频次分析方法计算得出,71%的长鳍金枪鱼中心渔场趋向于集中分布在20 ℃等温线深度为209~245 m(227±18)的海域,70%的长鳍金枪鱼中心渔场趋向于集中分布在25 ℃等温线深度为88~142 m(115±27)的海域。
采用K-S检验方法对长鳍金枪鱼高值CPUE与20 ℃和25 ℃等温线深度之间的关系进行检验,样本数为485,在给定显著性水平α=0.05下,得出D0.05=0.061 7。ECDF分析结果见图 5。20 ℃和25 ℃等温线深度对应的最大D(t)值分别为0.026 9和0.025 1,均远小于D0.05,表明样本间服从同一分布,长鳍金枪鱼高值CPUE与20 ℃和25 ℃等温线深度间有密切关系。20 ℃和25 ℃等温线深度对应的最大D(t)值分别出现在220 m和111 m,与中心渔场20 ℃和25 ℃等温线深度的均值稍有差异(分别为227 m和115 m)。采用ECDF方法得出长鳍金枪鱼中心渔场20 ℃和25 ℃等温线深度分别为202~238 m(220±18)、84~138 m(111±27)。综合频次分析和ECDF分析的结果,取两者交集,得出南太平洋长鳍金枪鱼适宜的垂直活动水层深度为88~238 m。
3. 讨论
3.1 长鳍金枪鱼渔场分布与20 ℃、25 ℃等温线深度的关系
以往对南太平洋长鳍金枪鱼渔场环境的研究,采用的多是遥感获取的海表面环境因子[15-19]。但标志放流和延绳钓调查结果表明,长鳍金枪鱼多分布于80 m以深的水层,10 m以浅的近表层很少有长鳍金枪鱼活动[20-21]。因此,相较于表层环境因子,采用次表层环境变量分析长鳍金枪鱼的栖息环境更为适合。长鳍金枪鱼耳石研究发现,长鳍金枪鱼经常出没于海洋环境变化较大的水层,如温跃层和混合层[22]。笔者在研究过程中也发现,15°S以南,长鳍金枪鱼高值CPUE密集分布的海域,其25 ℃等温线深度的等深线分布也十分密集,推测此处可能有混合层存在(图 3)。以往的研究中,常用20 ℃等温线深度来表示太平洋海域温跃层的深度,表明在太平洋海域次表层20 ℃等温线分布与温跃层的分布十分相似[23-24]。DOMOKOS等[6]指出,长鳍金枪鱼白天下潜至温跃层以下觅食DSL生物,夜晚则游回混合层上方,混合层及温跃层的深度直接影响到长鳍金枪鱼垂直活动水层的深度。对太平洋海域DSL生物的调查研究发现,DSL生物白天主要分布于温跃层以下,夜晚则分布于温跃层以上[25]。据此推断,20 ℃等温线可能通过影响DSL生物的垂直分布,进而影响长鳍金枪鱼垂直活动水层的空间分布。
K-S检验结果表明,长鳍金枪鱼高值CPUE与20 ℃和25 ℃等温线深度之间有密切关系。长鳍金枪鱼中心渔场所处海域,其20 ℃等温线深度多在220 m以深,超过250 m则少有中心渔场出现;25 ℃等温线深度多在140 m以浅,浅于80 m则少有中心渔场出现。长鳍金枪鱼高值CPUE与20 ℃和25 ℃等温线深度的时空分布均呈现出明显的季节性变化特征,20 ℃和25 ℃等温线深度的这种季节性变化进而导致长鳍金枪鱼垂直活动水层深度的季节性变化。长鳍金枪鱼中心渔场的时空分布与20 ℃等温线深度的220 m等深线和25 ℃等温线深度的140 m等深线的时空变动具有显著的相关性,进一步表明水温垂直结构对长鳍金枪鱼的垂直活动水层深度及渔场分布有直接影响。
3.2 长鳍金枪鱼适宜垂直活动水层深度分析
综合频次分析和ECDF方法计算得出,南太平洋长鳍金枪鱼适宜的垂直活动水层深度为88~238 m。所得结果在中西太平洋长鳍金枪鱼垂直分布水层(80~380 m)之内[19],与临近的所罗门群岛附近海域长鳍金枪鱼活动较为频繁的水层(130~190 m)相符[7],与相似纬度的东太平洋海域成年长鳍金枪鱼钓获深度(102.3~236.1 m)基本相同[26],与东南太平洋长鳍金枪鱼延绳钓钓获深度(74.33~296.50 m)相似[27]。文章初步得出了南太平洋长鳍金枪鱼适宜的垂直活动水层深度范围,在实际的长鳍金枪鱼延绳钓生产作业中,应尽可能使延绳钓钓钩沉降至88~238 m,从而有效节省饵料投放、提高长鳍金枪鱼的生产效率。
4. 展望
文章从次表层水温垂直结构的角度研究了南太平洋长鳍金枪鱼的垂直活动水层分布特征及其适宜的垂直活动水层深度范围,事实上,长鳍金枪鱼的垂直分布还受到海流、溶解氧、海水盐度、饵料生物分布等其他诸多环境因素的共同影响[28-30]。Argo数据可以实时获取次表层水温、水深、盐度以及溶解氧等信息,极大地拓展了远洋渔业资源和渔场学研究中海洋环境因子的选择范围,在远洋渔业研究中有着良好的应用前景。综合多种环境因子,恰当地选择神经网络、支持向量机、栖息地适应性指数等预报模型,准确、全面地把握渔场时空分布,是今后中国长鳍金枪鱼渔业研究的一个重要方向。
-
表 1 PCR反应体系的因素水平
Table 1 Factors and levels of PCR reaction
因素
factors水平(体系终浓度)/levels(final concentration) 1 2 3 4 Mg2+/mmol·L-1 1.60 2.00 2.20 2.40 dNTPs/mmol·L-1 0.10 0.20 0.30 0.40 TaqDNA聚合酶/U TaqDNA polymerase 0.50 1.00 1.50 2.00 引物/μmol·L-1 primer 0.10 0.20 0.30 0.40 模板/ng template 10 30 50 70 表 2 PCR反应的因素水平正交试验设计
Table 2 Factors and levels of PCR reaction based on orthogonal experimental design
编号
No.Mg2+
/mmol·L-1dNTPs
/mmol·L-1TaqDNA聚合酶/U
TaqDNA polymerase引物/μmol·L-1
primer模板/ng
template得分score 平均得分
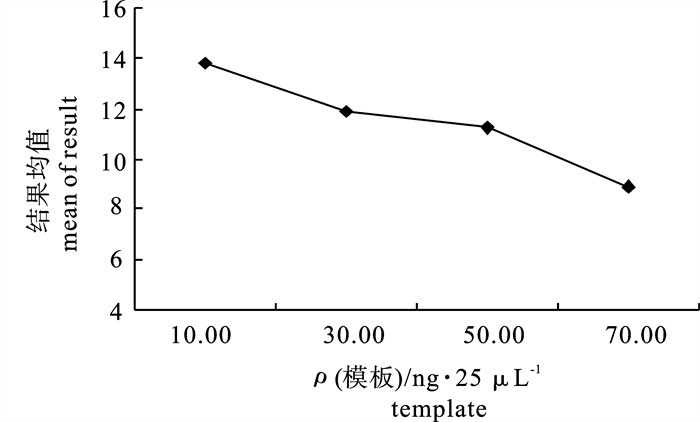
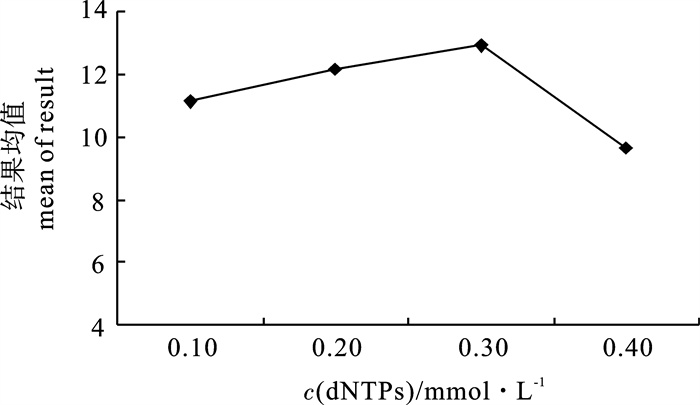
average score1 2 3 1 1.60 0.10 0.50 0.10 10 11 10 10 10.33 2 1.60 0.20 1.00 0.20 30 13 12 11 12.00 3 1.60 0.30 1.50 0.30 50 14 11 10 11.67 4 1.60 0.40 2.00 0.40 70 2 1 2 1.67 5 2.00 0.10 1.00 0.30 70 10 9 9 9.33 6 2.00 0.20 0.50 0.40 50 10 7 14 10.33 7 2.00 0.30 2.00 0.10 30 11 14 10 11.67 8 2.00 0.40 1.50 0.20 10 16 15 14 15.00 9 2.20 0.10 1.50 0.40 30 13 15 13 13.67 10 2.20 0.20 2.00 0.30 10 15 16 14 15.00 11 2.20 0.30 0.50 0.20 70 15 12 13 13.33 12 2.20 0.40 1.00 0.10 50 12 11 12 11.67 13 2.40 0.10 2.00 0.20 50 11 12 11 11.33 14 2.40 0.20 1.50 0.10 70 10 12 12 11.33 15 2.40 0.30 1.00 0.40 10 13 16 16 15.00 16 2.40 0.40 0.50 0.30 30 11 10 10 10.33 T1 35.67 44.67 44.33 45.00 55.33 - - - - T2 46.33 48.67 48.00 51.67 47.67 - - - - T3 53.67 51.67 51.67 46.33 45.00 - - - - T4 48.00 38.67 39.67 40.67 35.67 - - - - t1 8.92 11.17 11.08 11.25 13.83 - - - - t2 11.58 12.17 12.00 12.92 11.92 - - - - t3 13.42 12.92 12.92 11.58 11.25 - - - - t4 12.00 9.67 9.92 10.17 8.92 - - - - R 4.50 3.25 3.00 2.75 4.91 - - - - 注:T1~T4为各水平的平均得分总和;t1~t4分别为T1,T2,T3和T4的平均值Note:T1~T4 represent the sum of average scores of each treatment; t1~t4 represent the mean of T1, T2, T3, T4. -
[1] 李鹏飞, 刘萍, 李健, 等. 莱州湾三疣梭子蟹的生化遗传分析[J]. 海洋水产研究, 2007, 28(2): 90-96. doi: 10.3969/j.issn.1000-7075.2007.02.014 [2] 余红卫, 朱冬发, 韩宝芹. 三疣梭子蟹不同组织同工酶的分析[J]. 动物学杂志, 2005, 40(1): 84-87. doi: 10.3969/j.issn.0250-3263.2005.01.014 [3] 朱冬发, 余红卫, 王春琳. 三疣梭子蟹个体发育早期的同工酶谱变化[J]. 水产学报, 2005, 29(6): 751-756. [4] 王国良, 金珊, 李政, 等. 三疣梭子蟹养殖群体同工酶的组织特异性及生化遗传分析[J]. 台湾海峡, 2005, 24(4): 474-480. doi: 10.3969/j.issn.1000-8160.2005.04.009 [5] 高保全, 刘萍, 李健, 等. 三疣梭子蟹野生群体同工酶的遗传多态性分析[J]. 水产学报, 2007, 31(1): 1-6. doi: 10.3321/j.issn:1000-0615.2007.01.001 [6] 陈淑吟, 吉红九, 丁亚平, 等. 吕泗渔场三疣梭子蟹自然群体同工酶与ISSR遗传多样性分析[J]. 上海水产大学学报, 2008, 17(4): 406-410. https://xueshu.baidu.com/usercenter/paper/show?paperid=96ae324a3f5db3bc37f4d01704b5e62a&site=xueshu_se [7] 王敏强, 崔志峰, 刘晓玲, 等. 2种三疣梭子蟹居群线粒体Cyt b和S-rRNA基因片段序列变异研究[J]. 烟台大学学报: 自然科学与工程版, 2008, 21(3): 191-197. doi: 10.3969/j.issn.1004-8820.2008.03.008 [8] 郭天慧, 孔晓瑜, 陈四清, 等. 三疣梭子蟹线粒体DNA 16S rRNA和COI基因片段序列的比较研究[J]. 中国海洋大学学报, 2004, 34(1): 22-28. doi: 10.3969/j.issn.1672-5174.2004.01.002 [9] 张姝, 李喜莲, 崔朝霞, 等. 线粒体基因片段在梭子蟹系统发育及物种鉴定中的应用[J]. 海洋科学, 2008, 32(4): 9-18. https://xueshu.baidu.com/usercenter/paper/show?paperid=d662fff511c2b719a004bd69a41f7a99&site=xueshu_se&hitarticle=1 [10] 冯冰冰, 李家乐, 牛东红, 等. 我国沿海三疣梭子蟹9个野生群体线粒体CR和COI片段比较分析[J]. 动物学杂志, 2008, 43(2): 28-36. doi: 10.3969/j.issn.0250-3263.2008.02.005 [11] 冯冰冰, 李家乐, 牛东红, 等. 我国四大海域三疣梭子蟹线粒体控制区基因片段序列比较分析[J]. 上海水产大学学报, 2008, 17(2): 134-139. [12] 金珊, 赵青松, 王春琳, 等. 梭子蟹科六种海产蟹的RAPD标记[J]. 动物学研究, 2004, 25(2): 172-176. doi: 10.3321/j.issn:0254-5853.2004.02.016 [13] 刘爽, 薛淑霞, 孙金生. 黄海和东海三疣梭子蟹(Portunus triuberbuculatus)的AFLP分析[J]. 海洋与湖沼, 2008, 39(2): 152-156. doi: 10.11693/hyhz20080224024 [14] LI G, QUIROS C F. Sequence-related amplified polymorphism (SRAP), a new marker system based on a simple PCR reaction: its application to mapping and gene tagging in Brassica[J]. Theor Appl Genet, 2001, 103(2): 455-461. doi: 10.1007/s001220100570
[15] LIN Zhongxu, ZHANG Xianlong, NIE Yichun. Construction of a genetic linkage map for cotton based on SRAP[J]. Chin Sci Bull, 2003, 48(19): 2063-2067. doi: 10.1360/03wc0193
[16] LIN Zhongxu, ZHANG Xianlong, NIE Yichun. Evaluation of application of a new molecular marker SRAP analysis of F2 segregation population and genetic diversity in cotton[J]. Acta Genet Sin, 2004, 31(6): 622-626. doi: 10.1088/1009-0630/6/5/011
[17] BUDAK H, SHEARMAN R C, PARMAKSIZ I. Molecular characterization of buffalograss germplasm using sequence-related amplified polymorphism markers[J]. Theor Appl Genet, 2004, 108(2): 328-334. doi: 10.1007/s00122-003-1428-4
[18] BUDAK H, SHEARMAN RC, PARMAKSIZ I, et al. Comparative analysis of seeded and vegetative biotype buffalograsses based on phylogenetic relationship using ISSRs, SSRs, RAPDs and SRAPs[J]. Theor Appl Genet, 2004, 109(2): 280-288. doi: 10.1007/s00122-004-1630-z
[19] FERRIOL M, PICO B, NUEZ F. Genetic diversity of a germplasm collection of Cucurbita pepo using SRAP and AFLP markers[J]. Theor Appl Genet, 2003, 107(2): 271-282. doi: 10.1007/s00122-003-1242-z
[20] FERRIOL M, PICO B, NUEZ F. Genetic diversity of some accessions of Cucurbita maxima from Spain using RAPD and SRAP markers[J]. Genet Resour Crop Evol, 2003, 50(3): 227-238. doi: 10.1023/A:1023502925766
[21] 王燕青, 季孔庶. 利用正交设计优化牡丹SRAP-PCR反应体系[J]. 分子植物育种, 2009, 7(1): 199-203. doi: 10.3969/j.issn.1672-416X.2009.01.033 [22] 谢运海, 夏德安, 姜静, 等. 利用正交设计优化水曲柳ISSR-PCR反应体系[J]. 分子植物育种, 2005, 3(3): 445-450. doi: 10.3969/j.issn.1672-416X.2005.03.022 [23] 柳李旺, 龚义勤, 黄浩, 等. 新型分子标记—SRAP与TRAP及其应用[J]. 遗传, 2004, 26(5): 777-778. doi: 10.3321/j.issn:0253-9772.2004.05.039 [24] 郭大龙, 罗正荣. 部分柿属植物SRAP-PCR反应体系的优化[J]. 果树学报, 2006, 23(10): 138-141. doi: 10.3969/j.issn.1009-9980.2006.01.034 -
期刊类型引用(3)
1. 刘明泰, 李杨, 刘小林, 许淑芬, 常亚青. 皱纹盘鲍SRAP反应体系的正交优化. 大连海洋大学学报. 2013(01): 12-16 .  百度学术
百度学术
2. 智艳平, 武喜艳, 安瑶瑶, 郜刚. 青枯菌SRAP-PCR体系的建立与优化. 山西师范大学学报(自然科学版). 2013(04): 69-72 .  百度学术
百度学术
3. 程长洪, 张敏莹, 刘凯, 徐东坡, 段金荣, 周彦锋, 施炜纲. 暗纹东方鲀SRAP-PCR体系的建立与优化. 浙江农业学报. 2011(06): 1084-1089 .  百度学术
百度学术
其他类型引用(0)




 下载:
下载:








 粤公网安备 44010502001741号
粤公网安备 44010502001741号
